Genome-wide coexpression dynamics: theory and application
- PMID: 12486219
- PMCID: PMC139237
- DOI: 10.1073/pnas.252466999
Genome-wide coexpression dynamics: theory and application
Abstract
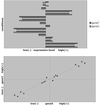
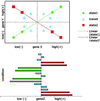
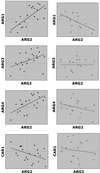
High-throughput expression profiling enables the global study of gene activities. Genes with positively correlated expression profiles are likely to encode functionally related proteins. However, all biological processes are interlocked, and each protein may play multiple cellular roles. Thus the coexpression of any two functionally related genes may depend on the constantly varying, yet often-unknown cellular state. To initiate a systematic study on this issue, a theory of coexpression dynamics is presented. This theory is used to rationalize a strategy of conducting a genome-wide search for the most critical cellular players that may affect the coexpression pattern of any two genes. In one example, using a yeast data set, our method reveals how the enzymes associated with the urea cycle are expressed to ensure proper mass flow of the involved metabolites. The correlation between ARG2 and CAR2 is found to change from positive to negative as the expression level of CPA2 increases. This delicate interplay in correlation signifies a remarkable control on the influx and efflux of ornithine and reflects well the intrinsic cellular demand for arginine. In addition to the urea cycle, our examples include SCH9 and CYR1 (both implicated in a recent longevity study), cytochrome c1 (mitochondrial electron transport), calmodulin (main calcium-binding protein), PFK1 and PFK2 (glycolysis), and two genes, ECM1 and YNL101W, the functions of which are newly revealed. The complexity in computation is eased by a new result from mathematical statistics.
Figures



References
-
- Scherf U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236-244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

