A polyphosphate kinase (PPK2) widely conserved in bacteria
- PMID: 12486232
- PMCID: PMC139203
- DOI: 10.1073/pnas.262655199
A polyphosphate kinase (PPK2) widely conserved in bacteria
Abstract
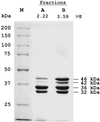
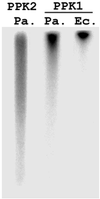
Synthesis of inorganic polyphosphate (poly P) from the terminal phosphate of ATP is catalyzed reversibly by poly P kinase (PPK, now designated PPK1) initially isolated from Escherichia coli. PPK1 is highly conserved in many bacteria, including some of the major pathogens such as Pseudomonas aeruginosa. In a null mutant of P. aeruginosa lacking ppk1, we have discovered a previously uncharacterized PPK activity (designated PPK2) distinguished from PPK1 by the following: synthesis of poly P from GTP or ATP, a preference for Mn2+ over Mg2+, and a stimulation by poly P. The reverse reaction, a poly P-driven nucleoside diphosphate kinase synthesis of GTP from GDP, is 75-fold greater than the forward reaction, poly P synthesis from GTP. The gene encoding PPK2 (ppk2) was identified from the amino acid sequence of the protein purified near 1,000-fold, to homogeneity. The 5'-end is 177 bp upstream of the annotated genome sequence of a "conserved hypothetical protein"; ppk2 (1,074 bp) encodes a protein of 357 aa with a molecular mass of 40.8 kDa. Sequences homologous to PPK2 are present in two other proteins in P. aeruginosa, in two Archaea, and in 32 other bacteria (almost all with PPK1 as well); these include rhizobia, cyanobacteria, Streptomyces, and several pathogenic species. Distinctive features of the poly P-driven nucleoside diphosphate kinase activity and structural aspects of PPK2 are among the subjects of an accompanying report.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

