Species-specific protein sequence and fold optimizations
- PMID: 12487631
- PMCID: PMC139977
- DOI: 10.1186/1471-2105-3-39
Species-specific protein sequence and fold optimizations
Abstract
Background: An organism's ability to adapt to its particular environmental niche is of fundamental importance to its survival and proliferation. In the largest study of its kind, we sought to identify and exploit the amino-acid signatures that make species-specific protein adaptation possible across 100 complete genomes.
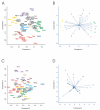
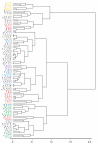
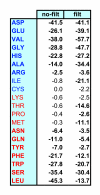
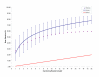
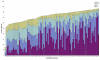
Results: Environmental niche was determined to be a significant factor in variability from correspondence analysis using the amino acid composition of over 360,000 predicted open reading frames (ORFs) from 17 archaea, 76 bacteria and 7 eukaryote complete genomes. Additionally, we found clusters of phylogenetically unrelated archaea and bacteria that share similar environments by amino acid composition clustering. Composition analyses of conservative, domain-based homology modeling suggested an enrichment of small hydrophobic residues Ala, Gly, Val and charged residues Asp, Glu, His and Arg across all genomes. However, larger aromatic residues Phe, Trp and Tyr are reduced in folds, and these results were not affected by low complexity biases. We derived two simple log-odds scoring functions from ORFs (CG) and folds (CF) for each of the complete genomes. CF achieved an average cross-validation success rate of 85 +/- 8% whereas the CG detected 73 +/- 9% species-specific sequences when competing against all other non-redundant CG. Continuously updated results are available at http://genome.mshri.on.ca.
Conclusion: Our analysis of amino acid compositions from the complete genomes provides stronger evidence for species-specific and environmental residue preferences in genomic sequences as well as in folds. Scoring functions derived from this work will be useful in future protein engineering experiments and possibly in identifying horizontal transfer events.
Figures



References
-
- Gross M, Jaenicke R. Proteins under pressure. The influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur J Biochem. 1994;221:617–30. - PubMed
-
- Audia JP, Webb CC, Foster JW. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol. 2001;291:97–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

