Characterization of genetic miscoding lesions caused by postmortem damage
- PMID: 12489042
- PMCID: PMC420012
- DOI: 10.1086/345379
Characterization of genetic miscoding lesions caused by postmortem damage
Abstract
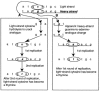
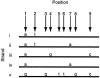
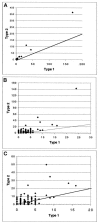
The spectrum of postmortem damage in mitochondrial DNA was analyzed in a large data set of cloned sequences from ancient human specimens. The most common forms of damage observed are two complementary groups of transitions, termed "type 1" (adenine-->guanine/thymine-->cytosine) and "type 2" (cytosine-->thymine/guanine-->adenine). Single-primer extension PCR and enzymatic digestion with uracil-N-glycosylase confirm that each of these groups of transitions result from a single event, the deamination of adenine to hypoxanthine, and cytosine to uracil, respectively. The predominant form of transition-manifested damage varies by sample, though a marked bias toward type 2 is observed with increasing amounts of damage. The two transition types can be used to identify the original strand, light (L) or heavy (H), on which the initial damage event occurred, and this can increase the number of detected jumping-PCR artifacts by up to 80%. No bias toward H-strand-specific damage events is noted within the hypervariable 1 region of human mitochondria, suggesting the rapid postmortem degradation of the secondary displacement (D-loop) H strand. The data also indicate that, as damage increases within a sample, fewer H strands retain the ability to act as templates for enzymatic amplification. Last, a significant correlation between archaeological site and sample-specific level of DNA damage was detected.
Figures
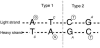

Similar articles
-
Postmortem miscoding lesions in sequence analysis of human ancient mitochondrial DNA.J Mol Evol. 2009 Jan;68(1):40-55. doi: 10.1007/s00239-008-9184-3. Epub 2008 Dec 6. J Mol Evol. 2009. PMID: 19067027
-
Assessing the fidelity of ancient DNA sequences amplified from nuclear genes.Genetics. 2006 Feb;172(2):733-41. doi: 10.1534/genetics.105.049718. Epub 2005 Nov 19. Genetics. 2006. PMID: 16299392 Free PMC article.
-
Recharacterization of ancient DNA miscoding lesions: insights in the era of sequencing-by-synthesis.Nucleic Acids Res. 2007;35(1):1-10. doi: 10.1093/nar/gkl483. Epub 2006 Aug 18. Nucleic Acids Res. 2007. PMID: 16920744 Free PMC article.
-
Base excision repair in nuclear and mitochondrial DNA.Prog Nucleic Acid Res Mol Biol. 2001;68:285-97. doi: 10.1016/s0079-6603(01)68107-8. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554304 Review.
-
Thymine DNA glycosylase.Prog Nucleic Acid Res Mol Biol. 2001;68:235-53. doi: 10.1016/s0079-6603(01)68103-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554300 Review.
Cited by
-
A "Copernican" reassessment of the human mitochondrial DNA tree from its root.Am J Hum Genet. 2012 Apr 6;90(4):675-84. doi: 10.1016/j.ajhg.2012.03.002. Am J Hum Genet. 2012. PMID: 22482806 Free PMC article.
-
High potential for using DNA from ancient herring bones to inform modern fisheries management and conservation.PLoS One. 2012;7(11):e51122. doi: 10.1371/journal.pone.0051122. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226474 Free PMC article.
-
Damage patterns observed in mtDNA control region MPS data for a range of template concentrations and when using different amplification approaches.Int J Legal Med. 2021 Jan;135(1):91-106. doi: 10.1007/s00414-020-02410-0. Epub 2020 Sep 17. Int J Legal Med. 2021. PMID: 32940843
-
Routine Mitogenome MPS Analysis from 1 and 5 mm of Rootless Human Hair.Genes (Basel). 2022 Nov 18;13(11):2144. doi: 10.3390/genes13112144. Genes (Basel). 2022. PMID: 36421819 Free PMC article.
-
Ancient DNA.Proc Biol Sci. 2005 Jan 7;272(1558):3-16. doi: 10.1098/rspb.2004.2813. Proc Biol Sci. 2005. PMID: 15875564 Free PMC article. Review.
References
-
- Anderson S, Bankier A, Arrell B, de Bruijn M, Coulson A, Drouin J, Eperon I, Nierlich D, Roe B, Sanger F, Schreier P, Smith A, Staden R, Young I (1981) Sequence and organisation of the human mitochondrial genome. Nature 290:457–465 - PubMed
-
- Ånensen H, Provan F, Lian A, Reinertsen S-H, Ueno Y, Matsuda A, Seeberg E, Bjelland S (2001) Mutations induced by 5-formyl-2′-deoxyuridine in Escherichia coli include base substitutions that can arise from mispairs of 5-formyluracil with guanine, cytosine and thymine. Mutat Res 476:99–107 - PubMed
-
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A (2002) Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295:2267–2270 - PubMed
-
- Burger J, Hummel S, Herrmann B, Henke W (1999) DNA preservation: a microsatellite-DNA study on ancient skeletal remains. Electophoresis 20:1722–1728 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

