Interaction of C5 protein with RNA aptamers selected by SELEX
- PMID: 12490703
- PMCID: PMC140078
- DOI: 10.1093/nar/gkf694
Interaction of C5 protein with RNA aptamers selected by SELEX
Abstract
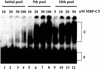
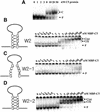
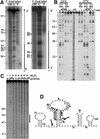
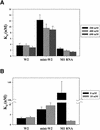
RNA aptamers binding to C5 protein, the protein component of Escherichia coli RNase P, were selected and characterized as an initial step in elucidating the mechanism of action of C5 protein as an RNA-binding protein. Sequence analyses of the RNA aptamers suggest that C5 protein binds various RNA molecules with dissociation constants comparable to that of M1 RNA, the RNA component of RNase P. The dominant sequence, W2, was chosen for further study. Interactions between W2 and C5 protein were independent of Mg2+, in contrast to the Mg2+ dependency of M1 RNA-C5 protein interactions. The affinity of W2 for C5 protein increased with increasing concentration of monovalent NH4+, suggesting interactions via hydrophobic attraction. W2 forms a fairly stable complex with C5 protein, although the stability of this complex is lower than that of the complex of M1 RNA with C5 protein. The core RNA motif essential for interaction with C5 protein was identified as a stem-loop structure, comprising a 5 bp stem and a 20 nt loop. Our results strongly imply that C5 protein is an interacting partner protein of some cellular RNA species apart from M1 RNA.
Figures



Similar articles
-
Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe.J Mol Biol. 2000 Feb 11;296(1):19-31. doi: 10.1006/jmbi.1999.3443. J Mol Biol. 2000. PMID: 10656815
-
Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli.Biochemistry. 1994 Feb 15;33(6):1399-405. doi: 10.1021/bi00172a016. Biochemistry. 1994. PMID: 8312258
-
Protein cofactor-dependent acquisition of novel catalytic activity by the RNase P ribonucleoprotein of E. coli.J Mol Biol. 2001 Apr 13;307(5):1181-212. doi: 10.1006/jmbi.2001.4519. J Mol Biol. 2001. PMID: 11292334
-
Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli.J Mol Biol. 1988 Aug 20;202(4):835-48. doi: 10.1016/0022-2836(88)90562-1. J Mol Biol. 1988. PMID: 2459398
-
From oligonucleotide shapes to genomic SELEX: novel biological regulatory loops.Proc Natl Acad Sci U S A. 1997 Jan 7;94(1):59-64. doi: 10.1073/pnas.94.1.59. Proc Natl Acad Sci U S A. 1997. PMID: 8990161 Free PMC article. Review.
Cited by
-
Complements are involved in alcoholic fatty liver disease, hepatitis and fibrosis.World J Hepatol. 2018 Oct 27;10(10):662-669. doi: 10.4254/wjh.v10.i10.662. World J Hepatol. 2018. PMID: 30386459 Free PMC article. Review.
-
Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing.Cell. 2018 Mar 22;173(1):181-195.e18. doi: 10.1016/j.cell.2018.02.034. Epub 2018 Mar 15. Cell. 2018. PMID: 29551268 Free PMC article.
-
GraphProt: modeling binding preferences of RNA-binding proteins.Genome Biol. 2014 Jan 22;15(1):R17. doi: 10.1186/gb-2014-15-1-r17. Genome Biol. 2014. PMID: 24451197 Free PMC article.
-
In vitro selection of RNA aptamers directed against protein E: a Haemophilus influenzae adhesin.Mol Biotechnol. 2014 Aug;56(8):714-25. doi: 10.1007/s12033-014-9749-x. Mol Biotechnol. 2014. PMID: 24682699
References
-
- Kurz J.C. and Fierke,C.A. (2000) Ribonuclease P: a ribonucleoprotein enzyme. Curr. Opin. Chem. Biol., 4, 553–558. - PubMed
-
- Frank D.N. and Pace,N.R. (1998) Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem., 67, 153–180. - PubMed
-
- Park B.H., Choi,Y.N., Park,J.W., Sim,S., Gil,M.C., Kim,S., Kim,M. and Lee,Y. (1998) Expression of C5 protein, the protein component of Escherichia coli RNase P, from the tac promoter. Mol. Cells, 8, 96–100. - PubMed
-
- Liu F. and Altman,S. (1994) Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell, 77, 1093–1100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

