Creation of a zymogen
- PMID: 12496934
- PMCID: PMC2819095
- DOI: 10.1038/nsb884
Creation of a zymogen
Abstract
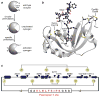
Cells produce proteases as inactive zymogens. Here, we demonstrate that this tactic can extend beyond proteases. By linking the N and C termini of ribonuclease A, we obstruct the active site with the amino acid sequence recognized by plasmepsin II, a highly specific protease from Plasmodium falciparum. We generate new N and C termini by circular permutation. In the presence of plasmepsin II, a ribonuclease zymogen gains approximately 10(3)-fold in catalytic activity and maintains high conformational stability. We conclude that zymogen creation provides a new and versatile strategy for the control of enzymatic activity, as well as the potential development of chemotherapeutic agents.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures


References
-
- Von Der Helm K, Korant BD, Cheronis JC, editors. Proteases as Targets for Therapy. Springer–Verlag; Heidelberg, Germany: 2000.
-
- Smith J, Simons C. Proteinase and Peptidase Inhibition: Recent Potential Targets for Drug Development. Taylor & Francis; London, UK: 2002.
-
- Kühne W. Ueber die Verdauung der Eiweissstoffe durch den Pankreassaft. Virchows Arch. 1867;39:130–174.
-
- Laskowski M, Jr, Qasim MA, Lu SM. In: Protein–Protein Recognition. Kleanthous C, editor. Oxford University Press; Oxford, UK: 2000. pp. 228–279.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
