VanD-type vancomycin-resistant Enterococcus faecium 10/96A
- PMID: 12499162
- PMCID: PMC149003
- DOI: 10.1128/AAC.47.1.7-18.2003
VanD-type vancomycin-resistant Enterococcus faecium 10/96A
Abstract
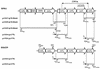
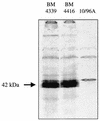
VanD type Enterococcus faecium 10/96A is constitutively resistant to vancomycin and to low levels of teicoplanin by nearly exclusive synthesis of peptidoglycan precursors terminating in D-alanyl-D-lactate (L. M. Dalla Costa, P. E. Reynolds, H. A. Souza, D. C. Souza, M. F. Palepou, and N. Woodford, Antimicrob. Agents Chemother. 44:3444-3446, 2000). A G(184)S mutation adjacent to the serine involved in the binding of D-Ala1 in the D-alanine:D-alanine ligase (Ddl) led to production of an impaired Ddl and accounts for the lack of D-alanyl-D-alanine-containing peptidoglycan precursors. The sequence of the vanD gene cluster revealed eight open reading frames. The organization of this operon, assigned to a chromosomal location, was similar to those in other VanD type strains. The distal part encoded the VanH(D) dehydrogenase, the VanD ligase, and the VanX(D) dipeptidase, which were homologous to the corresponding proteins in VanD-type strains. Upstream from the structural genes for these proteins was the vanY(D) gene; a frameshift mutation in this gene resulted in premature termination of the encoded protein and accounted for the lack of penicillin-susceptible D,D-carboxypeptidase activity. Analysis of the translated sequence downstream from the stop codon, but in a different reading frame because of the frameshift mutation, indicated homology with penicillin binding proteins (PBPs) with a high degree of identity with VanY(D) from VanD-type strains. The 5' end of the gene cluster contained the vanR(D)-vanS(D) genes for a putative two-component regulatory system. Insertion of ISEfa4 in the vanS(D) gene led to constitutive expression of vancomycin resistance. This new insertion belonged to the IS605 family and was composed of two open reading frames encoding putative transposases of two unrelated insertion sequence elements, IS200 and IS1341.
Figures




References
-
- Aiba, H., F. Nakasai, S. Mizushima, and T. Mizuno. 1989. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, results in stimulation of its DNA-binding ability. J. Biochem. 106:5-7. - PubMed
-
- Arthur, M., F. Depardieu, L. Cabanié, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX D,D-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. - PubMed
-
- Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and non partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849-1858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

