In34, a complex In5 family class 1 integron containing orf513 and dfrA10
- PMID: 12499211
- PMCID: PMC149023
- DOI: 10.1128/AAC.47.1.342-349.2003
In34, a complex In5 family class 1 integron containing orf513 and dfrA10
Abstract
A complex class 1 integron, In34, found in a conjugative plasmid from a multidrug-resistant Klebsiella pneumoniae strain isolated in 1997 at a hospital in Sydney, Australia, was shown to have a backbone related to that of In2, which belongs to the In5 family. In In34, the aadB gene cassette replaces the aadA1a cassette in In2, and two additional resistance genes, dfrA10 and aphA1, that are not part of a gene cassette are present. The aphA1 gene is in a Tn4352-like transposon that is located in the tniA gene. The dfrA10 gene lies adjacent to a 2,154-bp DNA segment, known as the common region, that contains an open reading frame predicting a product of 513 amino acids (Orf513). Orf513 is 66 and 55% identical to the products of two further open reading frames that, like the common region, are found adjacent to antibiotic resistance genes. A 27-bp conserved sequence was found at one end of each type of common region. The loss of dfrA10 due to homologous recombination between flanking direct repeats and incorporation of the excised circle by homologous recombination were demonstrated. Part of In34 is identical to the sequenced portion of In7, which is from a multidrug-resistant Escherichia coli strain that had been isolated 19 years earlier in the same hospital. In34 and In7 are in plasmids that contain the same six resistance genes conferring resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, neomycin, tobramycin, trimethoprim, and sulfonamides, but the plasmid backbones appear to be unrelated, suggesting that translocation of a multiple-drug-resistance-determining region as well as horizontal transfer may have occurred.
Figures
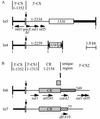



References
-
- Adrian, P. V., K. P. Klugman, and S. G. B. Aymes. 1995. Prevalence of trimethoprim resistant dihydrofolate reductase genes identified with oligonucleotide probes in plasmids from isolates of commensal faecal flora. J. Antimicrob. Chemother. 35:497-508. - PubMed
-
- Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

