Receptor-mediated endoproteolytic activation of two transcription factors in yeast
- PMID: 12502738
- PMCID: PMC187503
- DOI: 10.1101/gad.239202
Receptor-mediated endoproteolytic activation of two transcription factors in yeast
Abstract
Yeast possess a plasma membrane sensor of external amino acids that functions as a ligand-activated receptor. This multimeric sensor, dubbed the SPS sensor, initiates signals that regulate the expression of genes required for proper amino acid uptake. Stp1p and Stp2p are transcription factors that bind to specific sequences within the promoters of SPS-sensor-regulated genes. These factors exhibit redundant and overlapping abilities to activate transcription. We have found that Stp1p and Stp2p are synthesized as latent cytoplasmic precursors. In response to extracellular amino acids, the SPS sensor induces the rapid endoproteolytic processing of Stp1p and Stp2p. The processing of Stp1p/Stp2p occurs independently of proteasome function and without the apparent involvement of additional components. The shorter forms of these transcription factors, lacking N-terminal inhibitory domains, are targeted to the nucleus, where they transactivate SPS-sensor target genes. These results define a completely unique and streamline metabolic control pathway that directly routes environmental signals initiated at the plasma membrane to transcriptional activation in the nucleus of yeast.
Figures



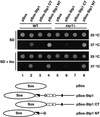



References
-
- André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. - PubMed
-
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
-
- Barnes D, Lai W, Breslav M, Naider F, Becker JM. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1998;29:297–310. - PubMed
-
- Bernard F, André B. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol Microbiol. 2001a;41:489–502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
