Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
- PMID: 12502741
- PMCID: PMC187509
- DOI: 10.1101/gad.1046102
Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
Abstract
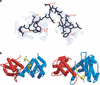
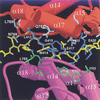
Repression of E2F transcription activity by the retinoblastoma (Rb) tumor suppressor through its interaction with the transactivation domain of the E2F transcription factor is one of the central features of G1/S arrest in the mammalian cell cycle. Deregulation of the Rb-E2F interaction results in hyperproliferation, lack of differentiation, and apoptosis, and can lead to cancer. The 2.2-A crystal structure of the Rb pocket complexed with an 18-residue transactivation-domain peptide of E2F-2 reveals that the boomerang-shaped peptide binds to the highly conserved interface between the A-box and the B-box of the Rb pocket in a bipartite manner. The N-terminal segment of the E2F-2 peptide in an extended beta-strand-like structure interacts with helices from the conserved groove at the A-B interface, whereas the C-terminal segment, which contains one 3(10) helix, binds to a groove mainly formed by A-box helices. The flexibility in the middle of the E2F-2 peptide is essential for the tight association of E2F to the Rb pocket. The binding of Rb to the E2F-2 peptide conceals several conserved residues that are crucial for transcription activation of E2F. We provide the structural basis for the Rb-mediated repression of E2F transcription activity without the requirement of histone-modifying enzymes.
Figures






Similar articles
-
A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites.Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11544-8. doi: 10.1073/pnas.92.25.11544. Proc Natl Acad Sci U S A. 1995. PMID: 8524800 Free PMC article.
-
Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release.Cell. 2005 Dec 16;123(6):1093-106. doi: 10.1016/j.cell.2005.09.044. Cell. 2005. PMID: 16360038
-
The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro.Nucleic Acids Res. 1993 Nov 11;21(22):4998-5004. doi: 10.1093/nar/21.22.4998. Nucleic Acids Res. 1993. PMID: 8255752 Free PMC article.
-
Small molecule regulators of Rb-E2F pathway as modulators of transcription.Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):788-94. doi: 10.1016/j.bbagrm.2010.07.004. Epub 2010 Jul 15. Biochim Biophys Acta. 2010. PMID: 20637913 Free PMC article. Review.
-
The RB/E2F pathway and regulation of RNA processing.Biochem Biophys Res Commun. 2009 Jul 3;384(3):280-3. doi: 10.1016/j.bbrc.2009.04.107. Epub 2009 May 3. Biochem Biophys Res Commun. 2009. PMID: 19401190 Free PMC article. Review.
Cited by
-
Structural Conservation and E2F Binding Specificity within the Retinoblastoma Pocket Protein Family.J Mol Biol. 2016 Oct 9;428(20):3960-3971. doi: 10.1016/j.jmb.2016.08.017. Epub 2016 Aug 25. J Mol Biol. 2016. PMID: 27567532 Free PMC article.
-
Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition.Mol Cell Biol. 2008 Jan;28(1):435-47. doi: 10.1128/MCB.00607-07. Epub 2007 Oct 29. Mol Cell Biol. 2008. PMID: 17967892 Free PMC article.
-
Small-molecule regulators that mimic transcription factors.Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):768-74. doi: 10.1016/j.bbagrm.2010.08.010. Epub 2010 Sep 6. Biochim Biophys Acta. 2010. PMID: 20804876 Free PMC article. Review.
-
PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation.Mol Cell Biol. 2015 Jan;35(1):238-48. doi: 10.1128/MCB.00945-14. Epub 2014 Oct 27. Mol Cell Biol. 2015. PMID: 25348716 Free PMC article.
-
Folding of a cyclin box: linking multitarget binding to marginal stability, oligomerization, and aggregation of the retinoblastoma tumor suppressor AB pocket domain.J Biol Chem. 2013 Jun 28;288(26):18923-38. doi: 10.1074/jbc.M113.467316. Epub 2013 Apr 30. J Biol Chem. 2013. PMID: 23632018 Free PMC article.
References
-
- Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-1, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. - PubMed
-
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. - PubMed
-
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
-
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstieve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: A new software suit for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
