ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase
- PMID: 12502744
- PMCID: PMC187497
- DOI: 10.1101/gad.239802
ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase
Abstract
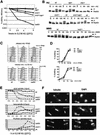
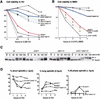
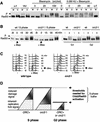
The intra-S-phase checkpoint in yeast responds to stalled replication forks by activating the ATM-like kinase Mec1 and the CHK2-related kinase Rad53, which in turn inhibit spindle elongation and late origin firing and lead to a stabilization of DNA polymerases at arrested forks. A mutation that destabilizes the second subunit of the Origin Recognition Complex, orc2-1, reduces the number of functional replication forks by 30% and severely compromises the activation of Rad53 by replication stress or DNA damage in S phase. We show that the restoration of the checkpoint response correlates in a dose-dependent manner with the restoration of pre-replication complex formation in G1. Other forms of DNA damage can compensate for the reduced level of fork-dependent signal in the orc2-1 mutant, yet even in wild-type cells, the amount of damage required for Rad53 activation is higher in S phase than in G2. Our data suggest the existence of an S-phase-specific threshold that may be necessary to allow cells to tolerate damage-like DNA structures present at normal replication forks.
Figures




References
-
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. - PubMed
-
- Bell SP. The origin recognition complex: From simple origins to complex functions. Genes & Dev. 2002;16:659–672. - PubMed
-
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. - PubMed
-
- Diffley JF. DNA replication: building the perfect switch. Curr Biol. 2001;11:R367–R370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
