Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin
- PMID: 12502807
- PMCID: PMC140850
- DOI: 10.1128/jvi.77.2.915-922.2003
Adenovirus type 5 DNA binding protein stimulates binding of DNA polymerase to the replication origin
Abstract
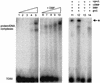
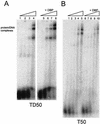
The adenovirus (Ad) DNA-binding protein (DBP) is essential for the elongation phase of Ad DNA replication by unwinding the template in an ATP-independent fashion, employing its capacity to form multimers. DBP also enhances the rate of initiation, with the highest levels obtained at low concentrations of Ad DNA polymerase (Pol). Here, we show that stimulation of initiation depends on the template conformation. Maximal stimulation, up to 15-fold, is observed on double-stranded or viral TP-containing origins. The stimulation is reduced on partially single-stranded origins and DBP does not enhance initiation any more once the origin is completely unwound. This suggests a role for DBP in origin unwinding that is comparable to its unwinding capacity during elongation. However, mutant DBP proteins defective in unwinding and elongation can still enhance initiation on ds templates. DBP also stimulates the binding of nuclear factor I (NFI) to the origin and lowers the K(m) for coupling of the first nucleotide to the precursor terminal protein by Pol. Mobility shift experiments reveal that DBP stimulates the binding of Pol on double-stranded origin and nonorigin DNA but not on single-stranded DNA. This effect is specific for DBP and is also seen with other DNA Pols. Our results suggest that, rather than by origin unwinding, DBP enhances initiation by modulating the origin conformation such that DNA Pol can bind more efficiently.
Figures

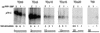



References
-
- Anderson, C. W., M. M. Hardy, J. J. Dunn, and D. F. Klessig. 1983. Independent, spontaneous mutants of adenovirus type 2-simian virus 40 hybrid Ad2+ND3 that grow efficiently in monkey cells possess identical mutations in the adenovirus type 2 DNA-binding protein gene. J. Virol. 48:31-39. - PMC - PubMed
-
- Ariga, H., H. Klein, A. J. Levine, and M. S. Horwitz. 1980. A cleavage product of the adenovirus DNA binding protein is active in DNA replication in vitro. Virology 101:307-310. - PubMed
-
- Bosher, J., I. R. Leith, S. M. Temperley, M. Wells, and R. T. Hay. 1991. The DNA-binding domain of nuclear factor I is sufficient to cooperate with the adenovirus type 2 DNA-binding protein in viral DNA replication. J. Gen. Virol. 72:2975-2980. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

