Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum
- PMID: 12502871
- PMCID: PMC140853
- DOI: 10.1128/jvi.77.2.1578-1583.2003
Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum
Abstract
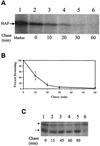
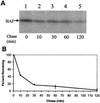
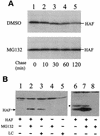
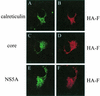
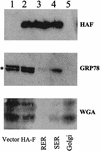
Hepatitis C virus (HCV) F protein is a newly discovered HCV gene product that is expressed by translational ribosomal frameshift. Little is known about the biological properties of this protein. By performing pulse-chase labeling experiments, we demonstrate here that the F protein is a labile protein with a half-life of <10 min in Huh7 hepatoma cells and in vitro. The half-life of the F protein could be substantially increased by proteasome inhibitors, suggesting that the rapid degradation of the F protein is mediated by the proteasome pathway. Further immunofluorescence staining and subcellular fractionation experiments indicate that the F protein is primarily associated with the endoplasmic reticulum. This subcellular localization is similar to those of HCV core and NS5A proteins, raising the possibility that the F protein may participate in HCV morphogenesis or replication.
Figures


Similar articles
-
Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus.J Virol. 2006 Mar;80(6):2832-41. doi: 10.1128/JVI.80.6.2832-2841.2006. J Virol. 2006. PMID: 16501092 Free PMC article.
-
Persistent and transient replication of full-length hepatitis C virus genomes in cell culture.J Virol. 2002 Apr;76(8):4008-21. doi: 10.1128/jvi.76.8.4008-4021.2002. J Virol. 2002. PMID: 11907240 Free PMC article.
-
The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein.Virology. 2001 May 25;284(1):70-81. doi: 10.1006/viro.2001.0873. Virology. 2001. PMID: 11352669
-
Signal peptide cleavage and internal targeting signals direct the hepatitis C virus p7 protein to distinct intracellular membranes.J Virol. 2005 Dec;79(24):15525-36. doi: 10.1128/JVI.79.24.15525-15536.2005. J Virol. 2005. PMID: 16306623 Free PMC article.
-
Differential subcellular localization of hepatitis C virus core gene products.Virology. 1995 Nov 10;213(2):455-61. doi: 10.1006/viro.1995.0018. Virology. 1995. PMID: 7491770
Cited by
-
Hepatitis C Virus Frameshift/Alternate Reading Frame Protein Suppresses Interferon Responses Mediated by Pattern Recognition Receptor Retinoic-Acid-Inducible Gene-I.PLoS One. 2016 Jul 12;11(7):e0158419. doi: 10.1371/journal.pone.0158419. eCollection 2016. PLoS One. 2016. PMID: 27404108 Free PMC article.
-
Toll-like Receptor Response to Hepatitis C Virus Infection: A Recent Overview.Int J Mol Sci. 2022 May 13;23(10):5475. doi: 10.3390/ijms23105475. Int J Mol Sci. 2022. PMID: 35628287 Free PMC article. Review.
-
Hepatitis C virus genetic variability and evolution.World J Hepatol. 2015 Apr 28;7(6):831-45. doi: 10.4254/wjh.v7.i6.831. World J Hepatol. 2015. PMID: 25937861 Free PMC article. Review.
-
Proapoptotic and pronecrosis effect of different truncated hepatitis C virus core proteins.J Zhejiang Univ Sci B. 2005 Apr;6(4):295-300. doi: 10.1631/jzus.2005.B0295. J Zhejiang Univ Sci B. 2005. PMID: 15754428 Free PMC article.
-
A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5' terminal sequence of the hepatitis C virus RNA.Mol Cell Proteomics. 2014 Jan;13(1):84-92. doi: 10.1074/mcp.M113.031682. Epub 2013 Oct 10. Mol Cell Proteomics. 2014. PMID: 24113282 Free PMC article.
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Blight, K. J., A. Grakoui, H. L. Hanson, and C. M. Rice. 2002. The molecular biology of hepatitis C virus, p. 81-108. In J.-H. J. Ou (ed.), Hepatitis viruses. Kluwer Academic Publishers, Norwell, Mass.
-
- Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

