Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase
- PMID: 12506195
- PMCID: PMC140885
- DOI: 10.1073/pnas.0135427100
Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase
Abstract
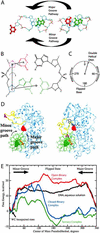
DNA methylation, various DNA repair mechanisms, and possibly early events in the opening of DNA as required for transcription and replication are initiated by flipping of a DNA base out of the DNA double helix. The energetics and structural mechanism of base flipping in the presence of the DNA-processing enzyme, cytosine 5-methyltransferase from HhaI (M.HhaI), were obtained through molecular dynamics based upon free-energy calculations. Free-energy profiles for base flipping show that, when in the closed conformation, M.HhaI lowers the free-energy barrier to flipping by 17 kcalmol and stabilizes the fully flipped state. Flipping is shown to occur via the major groove of the DNA. Structural analysis indicates that flipping is facilitated by destabilization of the DNA double-helical structure and substitution of DNA base-pairing and base-stacking interactions with DNA-protein interactions. The fully flipped state is stabilized by DNA-protein interactions that are enhanced upon binding of coenzyme. This study represents an atomic detail description of the mechanism by which a protein facilitates specific structural distortion in DNA.
Figures

Similar articles
-
Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase.Nucleic Acids Res. 2004 Jul 23;32(13):3877-86. doi: 10.1093/nar/gkh701. Print 2004. Nucleic Acids Res. 2004. PMID: 15273274 Free PMC article.
-
Atomistic view of base flipping in DNA.Philos Trans A Math Phys Eng Sci. 2004 Jul 15;362(1820):1439-60. doi: 10.1098/rsta.2004.1383. Philos Trans A Math Phys Eng Sci. 2004. PMID: 15306460 Review.
-
Spontaneous base flipping in DNA and its possible role in methyltransferase binding.Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000 Jul;62(1 Pt B):1133-7. doi: 10.1103/physreve.62.1133. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000. PMID: 11088571
-
The role of Arg165 towards base flipping, base stabilization and catalysis in M.HhaI.J Mol Biol. 2006 Sep 22;362(3):516-27. doi: 10.1016/j.jmb.2006.07.030. Epub 2006 Jul 22. J Mol Biol. 2006. PMID: 16926025
-
Structure, function, and mechanism of HhaI DNA methyltransferases.Crit Rev Biochem Mol Biol. 2002;37(3):167-97. doi: 10.1080/10409230290771492. Crit Rev Biochem Mol Biol. 2002. PMID: 12139442 Review.
Cited by
-
A molecular dynamics study of slow base flipping in DNA using conformational flooding.Biophys J. 2007 Aug 1;93(3):770-86. doi: 10.1529/biophysj.106.091751. Epub 2007 May 11. Biophys J. 2007. PMID: 17496048 Free PMC article.
-
Conformational transitions in RNA single uridine and adenosine bulge structures: a molecular dynamics free energy simulation study.Biophys J. 2006 Apr 1;90(7):2450-62. doi: 10.1529/biophysj.105.076158. Epub 2006 Jan 6. Biophys J. 2006. PMID: 16399833 Free PMC article.
-
Base-flipping mechanism in postmismatch recognition by MutS.Biophys J. 2011 Nov 2;101(9):2223-31. doi: 10.1016/j.bpj.2011.09.045. Epub 2011 Nov 1. Biophys J. 2011. PMID: 22067162 Free PMC article.
-
NMR imino proton exchange experiments on duplex DNA primarily monitor the opening of purine bases.J Am Chem Soc. 2006 Jan 25;128(3):678-9. doi: 10.1021/ja056445a. J Am Chem Soc. 2006. PMID: 16417331 Free PMC article.
-
Energetics of base flipping at a DNA mismatch site confined at the latch constriction of α-hemolysin.Faraday Discuss. 2016 Dec 12;193:471-485. doi: 10.1039/c6fd00058d. Faraday Discuss. 2016. PMID: 27711888 Free PMC article.
References
-
- Nakao M. Gene. 2001;278:25–31. - PubMed
-
- Roberts R J, Halford S S. In: Nucleases. Linn S M, Lloyd R S, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993.
-
- Plass C, Soloway P D. Eur J Hum Genet. 2002;10:6–16. - PubMed
-
- Goodman J I, Watson R E. Annu. Rev. Pharmacol. Toxicol. 2002. - PubMed
-
- Esteller M, Herman J G. J Pathol. 2002;196:1–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

