Positive and negative regulation of APP amyloidogenesis by sumoylation
- PMID: 12506199
- PMCID: PMC140945
- DOI: 10.1073/pnas.0235361100
Positive and negative regulation of APP amyloidogenesis by sumoylation
Erratum in
- Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):9102
Abstract
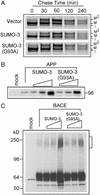
Amyloid beta peptide (Abeta) generated from amyloid precursor protein (APP) is central to Alzheimer's disease (AD). Signaling pathways affecting APP amyloidogenesis play critical roles in AD pathogenesis and can be exploited for therapeutic intervention. Here, we show that sumoylation, covalent modification of cellular proteins by small ubiquitin-like modifier (SUMO) proteins, regulates Abeta generation. Increased protein sumoylation resulting from overexpression of SUMO-3 dramatically reduces Abeta production. Conversely, reducing endogenous protein sumoylation with dominant-negative SUMO-3 mutants significantly increases Abeta production. We also show that mutant SUMO-3, K11R, which can only be monomerically conjugated to target proteins, has an opposite effect on Abeta generation to that by SUMO-3, which can form polymeric chains on target proteins. In addition, SUMO-3 immunoreactivity is predominantly detected in neurons in brains from AD, Down's syndrome, and nondemented humans. Therefore, polysumoylation reduces whereas monosumoylation or undersumoylation enhances Abeta generation. These findings provide a regulatory mechanism in APP amyloidogenesis and suggest that components in the sumoylation pathway may be critical in AD onset or progression.
Figures


Similar articles
-
Modulation of Abeta generation by small ubiquitin-like modifiers does not require conjugation to target proteins.Biochem J. 2007 Jun 1;404(2):309-16. doi: 10.1042/BJ20061451. Biochem J. 2007. PMID: 17346237 Free PMC article.
-
Assessing the Role of Paralog-Specific Sumoylation of HDAC1.Methods Mol Biol. 2017;1510:329-337. doi: 10.1007/978-1-4939-6527-4_24. Methods Mol Biol. 2017. PMID: 27761832
-
SUMO-1 modification increases human SOD1 stability and aggregation.Biochem Biophys Res Commun. 2006 Aug 25;347(2):406-12. doi: 10.1016/j.bbrc.2006.06.092. Epub 2006 Jun 23. Biochem Biophys Res Commun. 2006. PMID: 16828461
-
SUMO and Alzheimer's disease.Neuromolecular Med. 2013 Dec;15(4):720-36. doi: 10.1007/s12017-013-8257-7. Epub 2013 Aug 25. Neuromolecular Med. 2013. PMID: 23979993 Free PMC article. Review.
-
SUMOylation in carcinogenesis.Cancer Lett. 2012 Mar 28;316(2):113-25. doi: 10.1016/j.canlet.2011.10.036. Epub 2011 Nov 2. Cancer Lett. 2012. PMID: 22138131 Review.
Cited by
-
SUMOylation-disrupting WAS mutation converts WASp from a transcriptional activator to a repressor of NF-κB response genes in T cells.Blood. 2015 Oct 1;126(14):1670-82. doi: 10.1182/blood-2015-05-646182. Epub 2015 Aug 10. Blood. 2015. PMID: 26261240 Free PMC article.
-
Protein SUMOylation in neuropathological conditions.Drug News Perspect. 2009 Jun;22(5):255-65. doi: 10.1358/dnp.2009.22.5.1378636. Drug News Perspect. 2009. PMID: 19609463 Free PMC article. Review.
-
The E3 Ligase PIAS1 Regulates p53 Sumoylation to Control Stress-Induced Apoptosis of Lens Epithelial Cells Through the Proapoptotic Regulator Bax.Front Cell Dev Biol. 2021 Jun 14;9:660494. doi: 10.3389/fcell.2021.660494. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34195189 Free PMC article.
-
Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction.Physiol Rev. 2014 Oct;94(4):1249-85. doi: 10.1152/physrev.00008.2014. Physiol Rev. 2014. PMID: 25287864 Free PMC article. Review.
-
Trisomy of human chromosome 21 enhances amyloid-β deposition independently of an extra copy of APP.Brain. 2018 Aug 1;141(8):2457-2474. doi: 10.1093/brain/awy159. Brain. 2018. PMID: 29945247 Free PMC article.
References
-
- Hussain I, Powell D, Howlett D R, Tew D G, Meek T D, Chapman C, Gloger I S, Murphy K E, Southan C D, Ryan D M, et al. Mol Cell Neurosci. 1999;14:419–427. - PubMed
-
- Sinha S, Anderson J P, Barbour R, Basi G S, Caccavello R, Davis D, Doan M, Dovey H F, Frigon N, Hong J, et al. Nature. 1999;402:537–540. - PubMed
-
- Vassar R, Bennett B D, Babu-Kahn S, Kahn S, Mendiaz E A, Denis P, Teplow D B, Ross S, Amarante P, Loeloff R, et al. Science. 1999;286:735–741. - PubMed
-
- Yan R, Bienkowski M J, Shuck M E, Miao H, Tory M C, Pauley A M, Brashier J R, Stratman N C, Mathews W R, Buhl A E, et al. Nature. 1999;402:533–537. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

