Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer's disease is associated with the apolipoprotein E-epsilon4 allele
- PMID: 12507914
- PMCID: PMC1851126
- DOI: 10.1016/s0002-9440(10)63822-9
Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer's disease is associated with the apolipoprotein E-epsilon4 allele
Abstract
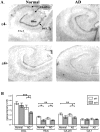
Abeta is the major component of amyloid plaques characterizing Alzheimer's disease (AD). Abeta accumulation can be affected by numerous factors including increased rates of production and/or impaired clearance. Insulin-degrading enzyme (IDE) has been implicated as a candidate enzyme responsible for the degradation and clearance of Abeta in the brain. We have previously shown that AD patients exhibit abnormalities in insulin metabolism that are associated with apoliprotein E (APOE) status. The possible association of IDE with AD, as well as the link between APOE status and insulin metabolism, led us to examine the expression of IDE in AD. We report that hippocampal IDE protein is reduced by approximately 50% in epsilon4+ AD patients compared to epsilon4- patients and controls. The allele-specific decrease of IDE in epsilon4+ AD patients is not associated with neuronal loss since neuron-specific enolase levels were comparable between the AD groups, regardless of APOE status. Hippocampal IDE mRNA levels were also reduced in AD patients with the epsilon4 allele compared to AD and normal subjects without the epsilon4 allele. These findings show that reduced IDE expression is associated with a significant risk factor for AD and suggest that IDE may interact with APOE status to affect Abeta metabolism.
Figures


References
-
- Haass C, Schlossmacher MG, Hung AY, Vigo-Pelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB, Selkoe DJ: Amyloid β-peptide is produced by cultured cells during normal metabolism. Nature 1992, 359:322-325 - PubMed
-
- Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, Cai XD, McKay DM, Tintner R, Frangione B, Younkin SG: Production of the Alzheimer amyloid β protein by normal proteolytic processing. Science 1992, 258:126-129 - PubMed
-
- Selkoe DJ: Clearing the brain’s amyloid cobwebs. Neuron 2001, 32:177-180 - PubMed
-
- Perez A, Morelli L, Cresto JC, Castano EM: Degradation of soluble amyloid β-peptides 1–40, 1–42, and the Dutch variant 1–40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem Res 2000, 25:247-255 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

