Alteration of mesodermal cell differentiation by EWS/FLI-1, the oncogene implicated in Ewing's sarcoma
- PMID: 12509448
- PMCID: PMC151529
- DOI: 10.1128/MCB.23.2.482-492.2003
Alteration of mesodermal cell differentiation by EWS/FLI-1, the oncogene implicated in Ewing's sarcoma
Abstract
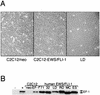
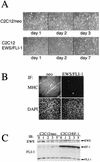
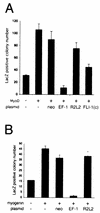
The chimeric fusion gene EWS/FLI-1 is detected in most cases of Ewing's sarcoma (ES), the second most common malignant bone tumor of childhood. Although 80% of ES tumors develop in skeletal sites, the remainder can arise in almost any soft tissue location. The lineage of the cell developing the EWS/FLI-1 gene fusion has not been fully characterized but is generally considered to be of either mesenchymal or neural crest origin. To study this oncogene in a conceptually relevant target cell, EWS/FLI-1 was introduced into the murine cell line C2C12, a myoblast cell line capable of differentiation into muscle, bone, or fat. In this cellular context, EWS/FLI-1 profoundly inhibited the myogenic differentiation program. The block in C2C12 myogenic differentiation required the nuclear localization and DNA-binding functions of EWS/FLI-1 and was mediated by transcriptional and posttranscriptional suppression of the myogenic transcription factors MyoD and myogenin. Interestingly, C2C12-EWS/FLI-1 cells constitutively expressed alkaline phosphatase, a bone lineage marker, and were alkaline phosphatase positive by histochemistry but showed no other evidence of bone lineage commitment. Consistent with recent findings in human ES tumor cell lines, C2C12-EWS/FLI-1 cells constitutively expressed cyclin D1 and demonstrated decreased expression of the cell cycle regulator p21(cip1), even under differentiation conditions and at confluent density. This C2C12-EWS/FLI-1 cell model may assist in the identification of novel differentially expressed genes relevant to ES and provide further insight into the cell(s) of origin developing ES-associated genetic fusions.
Figures






Similar articles
-
EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma.PLoS One. 2008 Apr 16;3(4):e1965. doi: 10.1371/journal.pone.0001965. PLoS One. 2008. PMID: 18414662 Free PMC article.
-
EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma.Cancer Res. 2008 Sep 1;68(17):7100-9. doi: 10.1158/0008-5472.CAN-07-6145. Cancer Res. 2008. PMID: 18757425 Free PMC article.
-
NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma.Mol Cancer Res. 2006 Nov;4(11):851-9. doi: 10.1158/1541-7786.MCR-06-0090. Mol Cancer Res. 2006. PMID: 17114343
-
EWS-FLI1 and Ewing's sarcoma: recent molecular data and new insights.Cancer Biol Ther. 2002 Jul-Aug;1(4):330-6. Cancer Biol Ther. 2002. PMID: 12432241 Review.
-
Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing's sarcoma.Cell Cycle. 2006 Sep;5(18):2049-53. doi: 10.4161/cc.5.18.3213. Epub 2006 Sep 15. Cell Cycle. 2006. PMID: 16969112 Review.
Cited by
-
The TET family of proteins: functions and roles in disease.J Mol Cell Biol. 2009 Dec;1(2):82-92. doi: 10.1093/jmcb/mjp025. Epub 2009 Sep 24. J Mol Cell Biol. 2009. PMID: 19783543 Free PMC article. Review.
-
Oncogenes, Proto-Oncogenes, and Lineage Restriction of Cancer Stem Cells.Int J Mol Sci. 2021 Sep 7;22(18):9667. doi: 10.3390/ijms22189667. Int J Mol Sci. 2021. PMID: 34575830 Free PMC article. Review.
-
Molecular basis of differentiation therapy for soft tissue sarcomas.Trends Cancer Res. 2010;6:69-90. Trends Cancer Res. 2010. PMID: 26912947 Free PMC article.
-
MYB oncoproteins: emerging players and potential therapeutic targets in human cancer.Oncogenesis. 2021 Feb 26;10(2):19. doi: 10.1038/s41389-021-00309-y. Oncogenesis. 2021. PMID: 33637673 Free PMC article. Review.
-
Inhibitor of DNA binding 2 (ID2) regulates the expression of developmental genes and tumorigenesis in ewing sarcoma.Oncogene. 2022 May;41(20):2873-2884. doi: 10.1038/s41388-022-02310-0. Epub 2022 Apr 14. Oncogene. 2022. PMID: 35422476 Free PMC article.
References
-
- Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene 20:5747-5754. - PubMed
-
- Bailly, R. A., R. Bosselut, J. Zucman, F. Cormier, O. Delattre, M. Roussel, G. Thomas, and J. Ghysdael. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 14:3230-3241. - PMC - PubMed
-
- Bennett, A. M., and N. K. Tonks. 1997. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science 278:1288-1291. - PubMed
-
- Buckingham, M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11:440-448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
