Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling
- PMID: 12509522
- PMCID: PMC143451
- DOI: 10.1105/tpc.006130
Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling
Abstract
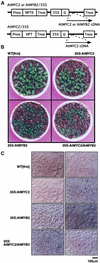
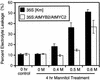
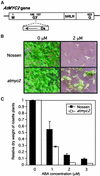
In Arabidopsis, the induction of a dehydration-responsive gene, rd22, is mediated by abscisic acid (ABA). We reported previously that MYC and MYB recognition sites in the rd22 promoter region function as cis-acting elements in the drought- and ABA-induced gene expression of rd22. bHLH- and MYB-related transcription factors, rd22BP1 (renamed AtMYC2) and AtMYB2, interact specifically with the MYC and MYB recognition sites, respectively, in vitro and activate the transcription of the beta-glucuronidase reporter gene driven by the MYC and MYB recognition sites in Arabidopsis leaf protoplasts. Here, we show that transgenic plants overexpressing AtMYC2 and/or AtMYB2 cDNAs have higher sensitivity to ABA. The ABA-induced gene expression of rd22 and AtADH1 was enhanced in these transgenic plants. Microarray analysis of the transgenic plants overexpressing both AtMYC2 and AtMYB2 cDNAs revealed that several ABA-inducible genes also are upregulated in the transgenic plants. By contrast, a Ds insertion mutant of the AtMYC2 gene was less sensitive to ABA and showed significantly decreased ABA-induced gene expression of rd22 and AtADH1. These results indicate that both AtMYC2 and AtMYB2 proteins function as transcriptional activators in ABA-inducible gene expression under drought stress in plants.
Figures





References
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199.
-
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54.
-
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

