Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection
- PMID: 12509524
- PMCID: PMC143454
- DOI: 10.1105/tpc.005165
Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection
Abstract
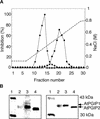
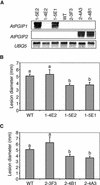
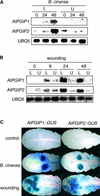
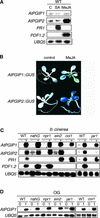
Polygalacturonase-inhibiting proteins (PGIPs) are plant proteins that counteract fungal polygalacturonases, which are important virulence factors. Like many other plant defense proteins, PGIPs are encoded by gene families, but the roles of individual genes in these families are poorly understood. Here, we show that in Arabidopsis, two tandemly duplicated PGIP genes are upregulated coordinately in response to Botrytis cinerea infection, but through separate signal transduction pathways. AtPGIP2 expression is mediated by jasmonate and requires COI1 and JAR1, whereas AtPGIP1 expression is upregulated strongly by oligogalacturonides but is unaffected by salicylic acid, jasmonate, or ethylene. Both AtPGIP1 and AtPGIP2 encode functional inhibitors of polygalacturonase from Botrytis, and their overexpression in Arabidopsis significantly reduces Botrytis disease symptoms. Therefore, gene duplication followed by the divergence of promoter regions may result in different modes of regulation of similar defensive proteins, thereby enhancing the likelihood of defense gene activation during pathogen infection.
Figures


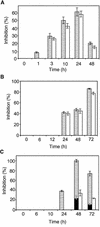
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Armand, S., Wagemaker, M.J., Sanchez-Torres, P., Kester H.C., van Santen, Y., Dijkstra, B.W., Visser, J., and Benen, J.A. (2000). The active site topology of Aspergillus niger endopolygalacturonase II as studied by site-directed mutagenesis. J. Biol. Chem. 275, 691–696. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2002). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates).
-
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

