Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA
- PMID: 12511486
- PMCID: PMC145342
- DOI: 10.1128/JB.185.2.413-421.2003
Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA
Abstract
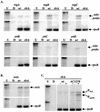
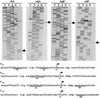
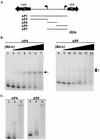
The proper temporal expression of virulence genes during infection is crucial to the infectious life cycle of microbial pathogens, particularly in pathogens that encounter a multitude of environments in eukaryotic hosts. Streptococcus pneumoniae normally colonizes the nasopharynges of healthy adults but can cause a range of diseases at a variety of host sites. Transcriptional regulators that are essential for full virulence of S. pneumoniae in different animal models have been identified. One such regulator, rlrA, is required for colonization of the nasopharynx and lung infection but is dispensable for systemic infection. Previous work has shown that rlrA lies in a 12-kb pathogenicity islet, divergently opposed to three putative sortase-anchored surface proteins and three sortase enzymes. In addition to rlrA, one of the putative surface proteins and one of the sortases have also been shown to be essential for lung infection. In this work, we demonstrate that RlrA is a positive regulator of all seven genes in the rlrA pathogenicity islet, with transcriptional activation occurring at four different promoters in the islet with AT-rich sequences. These promoters direct the expression of rlrA itself, the three sortases, rrgA, and rrgBC. These data are consistent with the model whereby the rlrA pathogenicity islet acts in an autonomous manner to alter the bacterial surface components that interact with the pulmonary and nasopharyngeal environments.
Figures

References
-
- Acebo, P., C. Nieto, M. A. Corrales, M. Espinosa, and P. Lopez. 2000. Quantitative detection of Streptococcus pneumoniae cells harbouring single or multiple copies of the gene encoding the green fluorescent protein. Microbiology 146:1267-1273. - PubMed
-
- Charpentier, E., and E. Tuomanen. 2000. Mechanisms of antibiotic resistance and tolerance in Streptococcus pneumoniae. Microbes Infect. 2:1855-1864. - PubMed
-
- Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. - PubMed
-
- Crook, D. W., and B. G. Spratt. 1998. Multiple antibiotic resistance in Streptococcus pneumoniae. Br. Med. Bull. 54:595-610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

