Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators
- PMID: 12511499
- PMCID: PMC145326
- DOI: 10.1128/JB.185.2.525-533.2003
Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators
Abstract
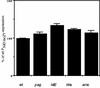
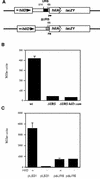
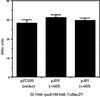
Salmonella enterica serovar Typhimurium causes human gastroenteritis and a systemic typhoid-like infection in mice. Infection is initiated by entry of the bacteria into intestinal epithelial cells and is mediated by a type III secretion system that is encoded by genes in Salmonella pathogenicity island 1. The expression of invasion genes is tightly regulated by environmental conditions such as oxygen and osmolarity, as well as by many bacterial factors. The hilA gene encodes an OmpR/ToxR family transcriptional regulator that activates the expression of invasion genes in response to both environmental and genetic regulatory factors. HilD is an AraC/XylS regulator that has been postulated to act as a derepressor of hilA expression that promotes transcription by interfering with repressor binding at the hilA promoter. Our research group has identified four genes (hilE, hha, pag, and ams) that negatively affect hilA transcription. Since the postulated function of HilD at the hilA promoter is to counteract the effects of repressors, we examined this model by measuring hilA::Tn5lacZY expression in strains containing negative regulator mutations in the presence or absence of functional HilD. Single negative regulator mutations caused significant derepression of hilA expression, and two or more negative regulator mutations led to very high level expression of hilA. However, in all strains tested, the absence of hilD resulted in low-level expression of hilA, suggesting that HilD is required for activation of hilA expression, whether or not negative regulators are present. We also observed that deletion of the HilD binding sites in the chromosomal hilA promoter severely decreased hilA expression. In addition, we found that a single point mutation at leucine 289 in the C-terminal domain of the alpha subunit of RNA polymerase leads to very low levels of hilA::Tn5lacZY expression, suggesting that HilD activates transcription of hilA by contacting and recruiting RNA polymerase to the hilA promoter.
Figures


References
-
- Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:1872-1882. - PubMed
-
- Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. - PubMed
-
- Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

