TmRNA is required for correct timing of DNA replication in Caulobacter crescentus
- PMID: 12511504
- PMCID: PMC145339
- DOI: 10.1128/JB.185.2.573-580.2003
TmRNA is required for correct timing of DNA replication in Caulobacter crescentus
Abstract
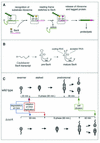
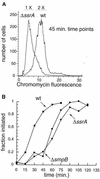
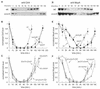
SsrA, or tmRNA, is a small RNA that interacts with selected translating ribosomes to target the nascent polypeptides for degradation. Here we report that SsrA activity is required for normal timing of the G(1)-to-S transition in Caulobacter crescentus. A deletion of the ssrA gene, or of the gene encoding SmpB, a protein required for SsrA activity, results in a specific delay in the cell cycle during the G(1)-to-S transition. The ssrA deletion phenotype is not due to accumulation of stalled ribosomes, because the deletion is not complemented by a mutated version of SsrA that releases ribosomes but does not target proteins for degradation. Degradation of the CtrA response regulator normally coincides with initiation of DNA replication, but in strains lacking SsrA activity there is a 40-min delay between the degradation of CtrA and replication initiation. This uncoupling of initiation of replication from CtrA degradation indicates that there is an SsrA-dependent pathway required for correct timing of DNA replication.
Figures

References
-
- Alley, M. R. K., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of the bacterial chemoreceptor for its targeted proteolysis. Science 259:1754-1757. - PubMed
-
- Barends, S., A. W. Karzai, R. T. Sauer, J. Wower, and B. Kraal. 2001. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 314:9-21. - PubMed
-
- Barends, S., J. Wower, and B. Kraal. 2000. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39:2652-2658. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

