ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression
- PMID: 12513977
- PMCID: PMC152388
- DOI: 10.1128/AEM.69.1.56-65.2003
ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression
Abstract
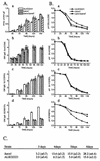
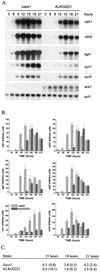
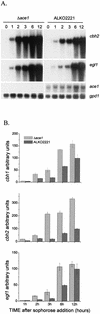
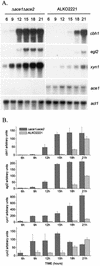
We characterized the effect of deletion of the Trichoderma reesei (Hypocrea jecorina) ace1 gene encoding the novel cellulase regulator ACEI that was isolated based on its ability to bind to and activate in vivo in Saccharomyces cerevisiae the promoter of the main cellulase gene, cbh1. Deletion of ace1 resulted in an increase in the expression of all the main cellulase genes and two xylanase genes in sophorose- and cellulose-induced cultures, indicating that ACEI acts as a repressor of cellulase and xylanase expression. Growth of the strain with a deletion of the ace1 gene on different carbon sources was analyzed. On cellulose-based medium, on which cellulases are needed for growth, the Deltaace1 strain grew better than the host strain due to the increased cellulase production. On culture media containing sorbitol as the sole carbon source, the growth of the strain with a deletion of the ace1 gene was severely impaired, suggesting that ACEI regulates expression of other genes in addition to cellulase and xylanase genes. A strain with a deletion of the ace1 gene and with a deletion of the ace2 gene coding for the cellulase and xylanase activator ACEII expressed cellulases and xylanases similar to the Deltaace1 strain, indicating that yet another activator regulating cellulase and xylanase promoters was present.
Figures

References
-
- Aro, N., A. Saloheimo, M. Ilmén, and M. Penttilä. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. - PubMed
-
- Bergmeyer, H. U. 1974. Methods of enzymatic analysis, 2nd ed., vol. 2. Academic Press, New York, N.Y.
-
- Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

