Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops
- PMID: 12514038
- PMCID: PMC152400
- DOI: 10.1128/AEM.69.1.533-541.2003
Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops
Abstract
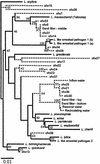
The total bacterial community of an experimental slow sand filter (SSF) was analyzed by denaturing gradient gel electrophoresis (DGGE) of partial 16S rRNA gene PCR products. One dominant band had sequence homology to Legionella species, indicating that these bacteria were a large component of the SSF bacterial community. Populations within experimental and commercial SSF units were studied by using Legionella-specific PCR primers, and products were studied by DGGE and quantitative PCR analyses. In the experimental SSF unit, the DGGE profiles for sand column, reservoir, storage tank, and headwater tank samples each contained at least one intense band, indicating that a single Legionella strain was predominant in each sample. Greater numbers of DGGE bands of equal intensity were detected in the outflow water sample. Sequence analysis of these PCR products showed that several Legionella species were present and that the organisms exhibited similarity to strains isolated from environmental and clinical samples. Quantitative PCR analysis of the SSF samples showed that from the headwater sample through the sand column, the number of Legionella cells decreased, resulting in a lower number of cells in the outflow water. In the commercial SSF, legionellae were also detected in the sand column samples. Storing prefilter water or locating SSF units within greenhouses, which are often maintained at temperatures that are higher than the ambient temperature, increases the risk of growth of Legionella and should be avoided. Care should also be taken when used filter sand is handled or replaced, and regular monitoring of outflow water would be useful, especially if the water is used for misting or overhead irrigation.
Figures



References
-
- Addiss, D. G., J. P. Davis, M. LaVenture, P. J. Wand, M. A. Hutchinson, and R. M. McKinney. 1989. Community-acquired Legionnaires' disease associated with a cooling tower: evidence for longer distance transport of Legionella pneumophila. Am. J. Epidemiol. 130:557-568. - PubMed
-
- Barletta, A. 1995. Isle of delight. Greenhouse Grower 9:28-31.
-
- Bender, L., M. Ott, R. Marre, and J. Hacker. 1990. Genome analysis of Legionella spp. by orthogonal field alternation gel electrophoresis (OFAGE). FEMS Microbiol. Lett. 60:253-257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

