Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein
- PMID: 12514740
- PMCID: PMC3727246
- DOI: 10.1038/nsb885
Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein
Abstract
The PutA flavoprotein from Escherichia coli plays multiple roles in proline catabolism by functioning as a membrane-associated bi-functional enzyme and a transcriptional repressor of proline utilization genes. The human homolog of the PutA proline dehydrogenase (PRODH) domain is critical in p53-mediated apoptosis and schizophrenia. Here we report the crystal structure of a 669-residue truncated form of PutA that shows both PRODH and DNA-binding activities, representing the first structure of a PutA protein and a PRODH enzyme from any organism. The structure is a domain-swapped dimer with each subunit comprising three domains: a helical dimerization arm, a 120-residue domain containing a three-helix bundle similar to that in the helix-turn-helix superfamily of DNA-binding proteins and a beta/alpha-barrel PRODH domain with a bound lactate inhibitor. Analysis of the structure provides insight into the mechanism of proline oxidation to pyrroline-5-carboxylate, and functional studies of a mutant protein suggest that the DNA-binding domain is located within the N-terminal 261 residues of E. coli PutA.
Figures

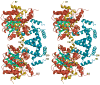
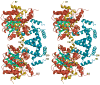
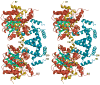






References
-
- Brown E, Wood JM. Redesigned Purification Yields a Fully functional PutA Protein Dimer from Escherichia coli. J Biol Chem. 1992;267:13086–13092. - PubMed
-
- Menzel R, Roth J. Purification of the putA gene Product. J Biol Chem. 1981;256:9755–9761. - PubMed
-
- Scarpulla RC, Soffer RL. Membrane-Bound Proline Dehydrogenase from Escherichia coli. J Biol Chem. 1978;253:5997–6001. - PubMed
-
- Menzel R, Roth J. Regulation of genes for Proline Utilization in Salmonella typhimurium: Autogenous Repression by the putA gene Product. J Mol Biol. 1981;148:21–44. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
