Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay
- PMID: 12517857
- PMCID: PMC149566
- DOI: 10.1128/JCM.41.1.254-260.2003
Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay
Abstract
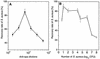
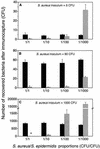
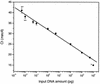
A rapid procedure was developed for detection and identification of methicillin-resistant Staphylococcus aureus (MRSA) directly from sterile sites or mixed flora samples (e.g., nose or inguinal swabs). After a rapid conditioning of samples, the method consists of two main steps: (i) immunomagnetic enrichment in S. aureus and (ii) amplification-detection profile on DNA extracts using multiplex quantitative PCR (5'-exonuclease qPCR, TaqMan). The triplex qPCR assay measures simultaneously the following targets: (i) mecA gene, conferring methicillin resistance, common to both S. aureus and Staphylococcus epidermidis; (ii) femA gene from S. aureus; and (iii) femA gene from S. epidermidis. This quantitative approach allows discrimination of the origin of the measured mecA signal. qPCR data were calibrated using two reference strains (MRSA and methicillin-resistant S. epidermidis) processed in parallel to clinical samples. This 96-well format assay allowed analysis of 30 swab samples per run and detection of the presence of MRSA with exquisite sensitivity compared to optimal culture-based techniques. The complete protocol may provide results in less than 6 h (while standard procedure needs 2 to 3 days), thus allowing prompt and cost-effective implementation of contact precautions.
Figures

References
-
- Abramson, M. A., and D. J. Sexton. 1999. Nosocomial methicillin-resistant and methicillin-susceptible Staphylococcus aureus primary bacteremia: at what costs? Infect. Control Hosp. Epidemiol. 20:408-411. - PubMed
-
- Alborn, W. E., Jr., J. Hoskins, S. Unal, J. E. Flokowitsch, C. A. Hayes, J. E. Dotzlaf, W. K. Yeh, and P. L. Skatrud. 1996. Cloning and characterization of femA and femB from Staphylococcus epidermidis. Gene 180:177-181. - PubMed
-
- Berger-Bachi, B., L. Barberis-Maino, A. Strassle, and F. H. Kayser. 1989. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 219:263-269. - PubMed
-
- Boyce, J. M. 2001. MRSA patients: proven methods to treat colonization and infection. J. Hosp. Infect. 48(Suppl. A):S9-S14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

