Crystal structure of saposin B reveals a dimeric shell for lipid binding
- PMID: 12518053
- PMCID: PMC140876
- DOI: 10.1073/pnas.0136947100
Crystal structure of saposin B reveals a dimeric shell for lipid binding
Abstract
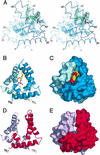
Saposin B is a small, nonenzymatic glycosphingolipid activator protein required for the breakdown of cerebroside sulfates (sulfatides) within the lysosome. The protein can extract target lipids from membranes, forming soluble protein-lipid complexes that are recognized by arylsulfatase A. The crystal structure of human saposin B reveals an unusual shell-like dimer consisting of a monolayer of alpha-helices enclosing a large hydrophobic cavity. Although the secondary structure of saposin B is similar to that of the known monomeric members of the saposin-like superfamily, the helices are repacked into a different tertiary arrangement to form the homodimer. A comparison of the two forms of the saposin B dimer suggests that extraction of target lipids from membranes involves a conformational change that facilitates access to the inner cavity.
Figures

References
-
- Sandhoff K, Kolter T. Trends Cell Biol. 1996;6:98–103. - PubMed
-
- Vaccaro A M, Salvioli R, Tatti M, Ciaffoni F. Neurochem Res. 1999;24:307–314. - PubMed
-
- Fluharty A L. Trends Glycosci Glycotechnol. 1995;7:167–189.
-
- Wilkening G, Linke T, Uhlhorn-Dierks G, Sandhoff K. J Biol Chem. 2000;275:35814–35819. - PubMed
-
- Munford R S, Sheppard P O, O'Hara P J. J Lipid Res. 1995;36:1653–1663. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

