MBGD: microbial genome database for comparative analysis
- PMID: 12519947
- PMCID: PMC165556
- DOI: 10.1093/nar/gkg109
MBGD: microbial genome database for comparative analysis
Abstract
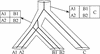
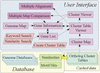
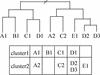
MBGD is a workbench system for comparative analysis of completely sequenced microbial genomes. The central function of MBGD is to create an orthologous gene classification table using precomputed all-against-all similarity relationships among genes in multiple genomes. In MBGD, an automated classification algorithm has been implemented so that users can create their own classification table by specifying a set of organisms and parameters. This feature is especially useful when the user's interest is focused on some taxonomically related organisms. The created classification table is stored into the database and can be explored combining with the data of individual genomes as well as similarity relationships among genomes. Using these data, users can carry out comparative analyses from various points of view, such as phylogenetic pattern analysis, gene order comparison and detailed gene structure comparison. MBGD is accessible at http://mbgd.genome.ad.jp/.
Figures


References
-
- Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

