STRING: a database of predicted functional associations between proteins
- PMID: 12519996
- PMCID: PMC165481
- DOI: 10.1093/nar/gkg034
STRING: a database of predicted functional associations between proteins
Abstract
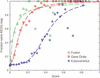
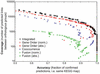
Functional links between proteins can often be inferred from genomic associations between the genes that encode them: groups of genes that are required for the same function tend to show similar species coverage, are often located in close proximity on the genome (in prokaryotes), and tend to be involved in gene-fusion events. The database STRING is a precomputed global resource for the exploration and analysis of these associations. Since the three types of evidence differ conceptually, and the number of predicted interactions is very large, it is essential to be able to assess and compare the significance of individual predictions. Thus, STRING contains a unique scoring-framework based on benchmarks of the different types of associations against a common reference set, integrated in a single confidence score per prediction. The graphical representation of the network of inferred, weighted protein interactions provides a high-level view of functional linkage, facilitating the analysis of modularity in biological processes. STRING is updated continuously, and currently contains 261 033 orthologs in 89 fully sequenced genomes. The database predicts functional interactions at an expected level of accuracy of at least 80% for more than half of the genes; it is online at http://www.bork.embl-heidelberg.de/STRING/.
Figures

References
-
- Galperin M.Y. and Koonin,E.V. (2000) Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. - PubMed
-
- Marcotte E.M. (2000) Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol., 10, 359–365. - PubMed
-
- Huynen M., Snel,B., Lathe,W. and Bork,P. (2000) Exploitation of gene context. Curr. Opin. Struct. Biol., 10, 366–370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

