Human immunodeficiency virus reverse transcriptase and protease sequence database
- PMID: 12520007
- PMCID: PMC165547
- DOI: 10.1093/nar/gkg100
Human immunodeficiency virus reverse transcriptase and protease sequence database
Abstract
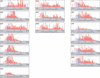
The HIV reverse transcriptase and protease sequence database is an on-line relational database that catalogues evolutionary and drug-related sequence variation in the human immunodeficiency virus (HIV) reverse transcriptase (RT) and protease enzymes, the molecular targets of antiretroviral therapy (http://hivdb.stanford.edu). The database contains a compilation of nearly all published HIV RT and protease sequences, including submissions to GenBank, sequences published in journal articles and sequences of HIV isolates from persons participating in clinical trials. Sequences are linked to data about the source of the sequence, the antiretroviral drug treatment history of the person from whom the sequence was obtained and the results of in vitro drug susceptibility testing. Sequence data on two new molecular targets of HIV drug therapy--gp41 (cell fusion) and integrase--will be added to the database in 2003.
Figures


References
-
- Hirsch M.S., Brun-Vezinet,F., D'Aquila,R.T., Hammer,S.M., Johnson,V.A., Kuritzkes,D.R., Loveday,C., Mellors,J.W., Clotet,B., Conway,B. et al. (2000) Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA, 283, 2417–2426. - PubMed
-
- Kilby J.M., Hopkins,S., Venetta,T.M., DiMassimo,B., Cloud,G.A., Lee,J.Y., Alldredge,L., Hunter,E., Lambert,D., Bolognesi,D. et al. (1998) Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nature Med., 4, 1302–1307. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

