Gene3D: structural assignments for the biologist and bioinformaticist alike
- PMID: 12520054
- PMCID: PMC165498
- DOI: 10.1093/nar/gkg051
Gene3D: structural assignments for the biologist and bioinformaticist alike
Abstract
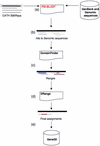
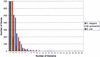
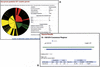
The Gene3D database (http://www.biochem.ucl.ac.uk/bsm/cath_new/Gene3D/) provides structural assignments for genes within complete genomes. These are available via the internet from either the World Wide Web or FTP. Assignments are made using PSI-BLAST and subsequently processed using the DRange protocol. The DRange protocol is an empirically benchmarked method for assessing the validity of structural assignments made using sequence searching methods where appropriate assignment statistics are collected and made available. Gene3D links assignments to their appropriate entries in relevent structural and classification resources (PDBsum, CATH database and the Dictionary of Homologous Superfamilies). Release 2.0 of Gene3D includes 62 genomes, 2 eukaryotes, 10 archaea and 40 bacteria. Currently, structural assignments can be made for between 30 and 40 percent of any given genome. In any genome, around half of those genes assigned a structural domain are assigned a single domain and the other half of the genes are assigned multiple structural domains. Gene3D is linked to the CATH database and is updated with each new update of CATH.
Figures

References
-
- Bray J., Todd,A., Pearl,F., Thornton,J. and Orengo,C. (2000) The CATH Dictionary of Homologous Superfamilies (DHS): a consensus approach for identifying distant structural homologues. Protein Eng., 13, 153–165. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

