MolMovDB: analysis and visualization of conformational change and structural flexibility
- PMID: 12520056
- PMCID: PMC165551
- DOI: 10.1093/nar/gkg104
MolMovDB: analysis and visualization of conformational change and structural flexibility
Abstract
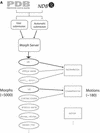
The Database of Macromolecular Movements (http://MolMovDB.org) is a collection of data and software pertaining to flexibility in protein and RNA structures. The database is organized into two parts. Firstly, a collection of 'morphs' of solved structures representing different states of a molecule provides quantitative data for flexibility and a number of graphical representations. Secondly, a classification of known motions according to type of conformational change (e.g. 'hinged domain' or 'allosteric') incorporates textual annotation and information from the literature relating to the motion, linking together many of the morphs. A variety of subsets of the morphs are being developed for use in statistical analyses. In particular, for each subset it is possible to derive distributions of various motional quantities (e.g. maximum rotation) that can be used to place a specific motion in context as being typical or atypical for a given population. Over the past year, the database has been greatly expanded and enhanced to incorporate new structures and to improve the quality of data. The 'morph server', which enables users of the database to add new morphs either from their own research or the PDB, has also been enhanced to handle nucleic acid structures and multi-chain complexes.
Figures


References
-
- Gerstein M., Lesk,A. and Chothia,C. (1994) Structural mechanisms for domain movements. Biochemistry, 33, 6739–6749. - PubMed
-
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. - PubMed
-
- Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH- A hierarchic classification of protein domain structures. Structure, 5, 1093–1108. - PubMed
-
- Qian J., Stenger,B., Wilson,C.A., Lin,J., Jansen,R., Teichmann,S.A., Park,J., Krebs,W.G., Yu,H., Alexandrov,V., Echols,N. and Gerstein,M. (2001) PartsList: a web-based system for dynamically ranking protein folds based on disparate attributes, including whole-genome expression and interaction information. Nucleic Acids Res., 29, 1750–1764. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

