RNABase: an annotated database of RNA structures
- PMID: 12520063
- PMCID: PMC165459
- DOI: 10.1093/nar/gkg012
RNABase: an annotated database of RNA structures
Abstract
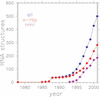
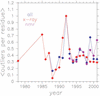
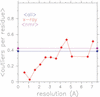
RNABase is a unified database of all three-dimensional structures containing RNA deposited in either the Protein Data Bank (PDB) or Nucleic Acid Data Base (NDB). For each structure, RNABase contains a brief summary as well as annotation of conformational parameters, identification of possible model errors, Ramachandran-style conformational maps and classification of ribonucleotides into conformers. These same analyses can also be performed on structures submitted by users. To facilitate access, structures are automatically placed into a variety of functional and structural categories, including: ribozymes, pseudoknots, etc. RNABase can be freely accessed on the web at http://www.rnabase.org. We are committed to maintaining this database indefinitely.
Figures