The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules
- PMID: 12522267
- PMCID: PMC141021
- DOI: 10.1073/pnas.212651999
The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules
Abstract
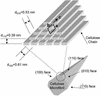
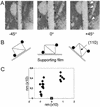
Cellulose binding modules (CBMs) potentiate the action of cellulolytic enzymes on insoluble substrates. Numerous studies have established that three aromatic residues on a CBM surface are needed for binding onto cellulose crystals and that tryptophans contribute to higher binding affinity than tyrosines. However, studies addressing the nature of CBM-cellulose interactions have so far failed to establish the binding site on cellulose crystals targeted by CBMs. In this study, the binding sites of CBMs on Valonia cellulose crystals have been visualized by transmission electron microscopy. Fusion of the CBMs with a modified staphylococcal protein A (ZZ-domain) allowed direct immuno-gold labeling at close proximity of the actual CBM binding site. The transmission electron microscopy images provide unequivocal evidence that the fungal family 1 CBMs as well as the family 3 CBM from Clostridium thermocellum CipA have defined binding sites on two opposite corners of Valonia cellulose crystals. In most samples these corners are worn to display significant area of the hydrophobic (110) plane, which thus constitutes the binding site for these CBMs.
Figures



References
-
- Linder M, Teeri T T. J Biotechnol. 1997;57:15–28. - PubMed
-
- Tomme P, Warren R A J, Miller R C, Kilburn D G, Gilkes N R. ACS Symp Ser. 1995;618:143–163.
-
- Reinikainen T, Teleman O, Teeri T T. Proteins. 1995;22:392–403. - PubMed
-
- Kraulis J, Clore G M, Nilges M, Jones T A, Pettersson G, Knowles J, Gronenborn A M. Biochemistry. 1989;28:7241–7257. - PubMed
-
- Mattinen M L, Linder M, Teleman A, Annila A. FEBS Lett. 1997;407:291–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

