Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio
- PMID: 12525633
- PMCID: PMC140984
- DOI: 10.1128/jvi.77.3.1992-2002.2003
Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio
Erratum in
- J Virol. 2003 Mar;77(6):3890.
Abstract
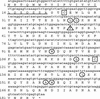
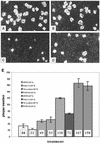
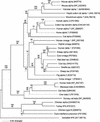
The interferon (IFN) family consisting of alpha IFN (IFN-alpha), IFN-beta, IFN-omega, IFN-delta, IFN-kappa, and IFN-tau is a large group of cytokines involved in the innate immune response against various microorganisms. Genes for IFN have been cloned from a variety of mammalian and avian species; however, IFN genes from lower-order vertebrates have not been forthcoming. Here, we report the cloning and characterization of the IFN gene from the zebrafish, Danio rerio. Zebrafish IFN (zfIFN) is 185 amino acids in length, with the first 22 amino acids representing a putative signal peptide. Treatment with the known IFN inducer polyinosinic acid-polycytidylic acid (poly[I]-poly[C]) resulted in an increase in zfIFN mRNA transcripts. zfIFN was also able to activate the IFN-inducible Mx promoter when cotransfected with a plasmid containing the zebrafish Mx promoter upstream of a luciferase reporter gene. To demonstrate antiviral activity, zebrafish cells were transfected with zfIFN and challenged with a fish rhabdovirus. A 36% reduction in plaque number was seen in zfIFN-transfected cells, compared to cells transfected with a control vector. Phylogenetic analysis has shown zfIFN to be approximately equally divergent from avian and mammalian IFN, consistent with its origin from an IFN present in the most recent common ancestor of these divergent lineages. A putative IFN from puffer, Fugu rubripes, was also found when zfIFN was used to search the fugu genome database, demonstrating that zfIFN can be used to find additional fish IFN genes. These results demonstrate that zebrafish can be used as an effective model for studying innate immunity and immune response to infectious disease.
Figures






References
-
- Berlocher, S. H., and D. L. Swofford. 1997. Searching for phylogenetic trees under the frequency parsimony criterion: an approximation using generalized parsimony. Syst. Biol. 46:211-215. - PubMed
-
- Chawla-Sarkar, M., D. W. Leaman, and E. C. Borden. 2001. Preferential induction of apoptosis by interferon (IFN)-β compared with IFN-α2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin. Cancer Res 7:1821-1831. - PubMed
-
- Collet, B., and C. J. Secombes. 2001. The rainbow trout (Oncorhynchus mykiss) Mx1 promoter. Structural and functional characterization. Eur. J. Biochem. 268:1577-1584. - PubMed
-
- Collodi, P., C. L. Miranda, X. Zhao, D. R. Buhler, and D. W. Barnes. 1994. Induction of zebrafish (Brachydanio rerio) P450 in vivo and in cell culture. Xenobiotica 24:487-493. - PubMed
-
- Daugherty, B., D. Martin-Zanca, B. Kelder, K. Collier, T. C. Seamans, K. Hotta, and S. Pestka. 1984. Isolation and bacterial expression of a murine alpha leukocyte interferon gene. J. Interferon Res. 4:635-643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

