Importance of basic residues in binding of rous sarcoma virus nucleocapsid to the RNA packaging signal
- PMID: 12525635
- PMCID: PMC140994
- DOI: 10.1128/jvi.77.3.2010-2020.2003
Importance of basic residues in binding of rous sarcoma virus nucleocapsid to the RNA packaging signal
Erratum in
- J Virol. 2003 Apr;77(7):4468
Abstract
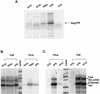
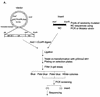
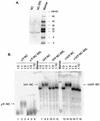
In the context of the Rous sarcoma virus Gag polyprotein, only the nucleocapsid (NC) domain is required to mediate the specificity of genomic RNA packaging. We have previously showed that the Saccharomyces cerevisiae three-hybrid system provides a rapid genetic assay to analyze the RNA and protein components of the avian retroviral RNA-Gag interactions necessary for specific encapsidation. In this study, using both site-directed mutagenesis and in vivo random screening in the yeast three-hybrid binding assay, we have examined the amino acids in NC required for genomic RNA binding. We found that we could delete either of the two Cys-His boxes without greatly abrogating either RNA binding or packaging, although the two Cys-His boxes are likely to be required for efficient viral assembly and release. In contrast, substitutions for the Zn-coordinating residues within the boxes did prevent RNA binding, suggesting changes in the overall conformation of the protein. In the basic region between the two Cys-His boxes, three positively charged residues, as well as basic residues flanking the two boxes, were necessary for both binding and packaging. Our results suggest that the stretches of positively charged residues within NC that need to be in a proper conformation appear to be responsible for selective recognition and binding to the packaging signal (Psi)-containing RNAs.
Figures






References
-
- Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

