Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection
- PMID: 12525648
- PMCID: PMC140883
- DOI: 10.1128/jvi.77.3.2134-2146.2003
Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection
Abstract
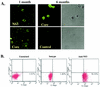
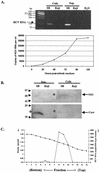
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Studies of HCV replication and pathogenesis have so far been hampered by the lack of an efficient tissue culture system for propagating HCV in vitro. Although HCV is primarily a hepatotropic virus, an increasing body of evidence suggests that HCV also replicates in extrahepatic tissues in natural infection. In this study, we established a B-cell line (SB) from an HCV-infected non-Hodgkin's B-cell lymphoma. HCV RNA and proteins were detectable by RNase protection assay and immunoblotting. The cell line continuously produces infectious HCV virions in culture. The virus particles produced from the culture had a buoyant density of 1.13 to 1.15 g/ml in sucrose and could infect primary human hepatocytes, peripheral blood mononuclear cells (PBMCs), and an established B-cell line (Raji cells) in vitro. The virus from SB cells belongs to genotype 2b. Single-stranded conformational polymorphism and sequence analysis of the viral RNA quasispecies indicated that the virus present in SB cells most likely originated from the patient's spleen and had an HCV RNA quasispecies pattern distinct from that in the serum. The virus production from the infected primary hepatocytes showed cyclic variations. In addition, we have succeeded in establishing several Epstein-Barr virus-immortalized B-cell lines from PBMCs of HCV-positive patients. Two of these cell lines are positive for HCV RNA as detected by reverse transcriptase PCR and for the nonstructural protein NS3 by immunofluorescence staining. These observations unequivocally establish that HCV infects B cells in vivo and in vitro. HCV-infected cell lines show significantly enhanced apoptosis. These B-cell lines provide a reproducible cell culture system for studying the complete replication cycle and biology of HCV infections.
Figures









References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Bouvier-Alias, M., K. Patel, H. Dahari, S. Beaucourt, P. Larderie, L. Blatt, C. Hezode, G. Picchio, D. Dhumeaux, A. U. Neumann, J. G. McHutchison, and J.-M. Pawlotsky. 2002. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211-218. - PubMed
-
- Cerny, A., and F. V. Chisari. 1999. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 30:595-601. - PubMed
-
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

