The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties
- PMID: 12525654
- PMCID: PMC140944
- DOI: 10.1128/jvi.77.3.2195-2206.2003
The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties
Abstract
Several studies have suggested the involvement of cutaneous human papillomaviruses (HPVs) in the development of nonmelanoma skin cancers. Here we have characterized the in vitro properties of E7 proteins of three cutaneous HPV types, 10, 20, and 38, which are frequently detected in skin specimens. We show that HPV38 E7 is able to inactivate the tumor suppressor pRb and induces loss of G(1)/S transition control, a key event in carcinogenesis. In contrast, HPV10 and HPV20 E7 proteins do not display these in vitro transforming activities. We also show that the two early proteins E6 and E7 of HPV38 are sufficient to corrupt the cell cycle and senescence programs in primary cells, inducing active and long-lasting proliferation of primary human keratinocytes, the natural host cells. Our study shows that E6 and E7 of this cutaneous HPV type have transforming activity in primary human cells, suggesting a role for HPV38 infection in skin carcinogenesis. In further support of such a role, we detected HPV38 DNA in approximately 50% of nonmelanoma skin cancers, but only in 10% of healthy skin specimens (P < 0.001).
Figures
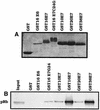


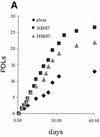











References
-
- Alunni-Fabbroni, M., T. Littlewood, L. Deleu, S. Caldeira, M. Giarre, M. Dell' Orco, and M. Tommasino. 2000. Induction of S phase and apoptosis by the human papillomavirus type 16 E7 protein are separable events in immortalized rodent fibroblasts. Oncogene 19:2277-2285. - PubMed
-
- Ananthaswamy, H. N., S. M. Loughlin, P. Cox, R. L. Evans, S. E. Ullrich, and M. L. Kripke. 1997. Sunlight and skin cancer: inhibition of p53 mutations in UV-irradiated mouse skin by sunscreens. Nat. Med. 3:510-514. - PubMed
-
- Ananthaswamy, H. N., and W. E. Pierceall. 1992. Molecular alterations in human skin tumors. Prog. Clin. Biol. Res. 376:61-84. - PubMed
-
- Armstrong, B. K., and A. Kricker. 2001. The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B 63:8-18. - PubMed
-
- Astori, G., D. Lavergne, C. Benton, B. Hockmayr, K. Egawa, C. Garbe, and E. M. de Villiers. 1998. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J. Investig. Dermatol. 110:752-755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

