Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development
- PMID: 12527753
- PMCID: PMC2172648
- DOI: 10.1083/jcb.200211046
Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development
Abstract
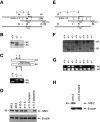
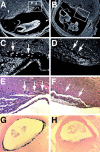
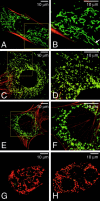
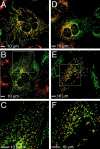
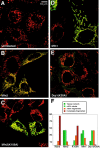
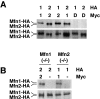
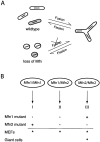
Mitochondrial morphology is determined by a dynamic equilibrium between organelle fusion and fission, but the significance of these processes in vertebrates is unknown. The mitofusins, Mfn1 and Mfn2, have been shown to affect mitochondrial morphology when overexpressed. We find that mice deficient in either Mfn1 or Mfn2 die in midgestation. However, whereas Mfn2 mutant embryos have a specific and severe disruption of the placental trophoblast giant cell layer, Mfn1-deficient giant cells are normal. Embryonic fibroblasts lacking Mfn1 or Mfn2 display distinct types of fragmented mitochondria, a phenotype we determine to be due to a severe reduction in mitochondrial fusion. Moreover, we find that Mfn1 and Mfn2 form homotypic and heterotypic complexes and show, by rescue of mutant cells, that the homotypic complexes are functional for fusion. We conclude that Mfn1 and Mfn2 have both redundant and distinct functions and act in three separate molecular complexes to promote mitochondrial fusion. Strikingly, a subset of mitochondria in mutant cells lose membrane potential. Therefore, mitochondrial fusion is essential for embryonic development, and by enabling cooperation between mitochondria, has protective effects on the mitochondrial population.
Figures



References
-
- Bakeeva, L.E., S. Chentsov Yu, and V.P. Skulachev. 1978. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim. Biophys. Acta. 501:349–369. - PubMed
-
- Bakeeva, L.E., Y.S. Chentsov, and V.P. Skulachev. 1981. Ontogenesis of mitochondrial reticulum in rat diaphragm muscle. Eur. J. Cell Biol. 25:175–181. - PubMed
-
- Bakeeva, L.E., S. Chentsov Yu, and V.P. Skulachev. 1983. Intermitochondrial contacts in myocardiocytes. J. Mol. Cell. Cardiol. 15:413–420. - PubMed
-
- Bereiter-Hahn, J., and M. Voth. 1994. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27:198–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

