Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution
- PMID: 12529302
- PMCID: PMC430971
- DOI: 10.1101/gr.844103
Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution
Abstract
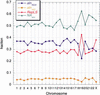
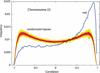
Six measures of evolutionary change in the human genome were studied, three derived from the aligned human and mouse genomes in conjunction with the Mouse Genome Sequencing Consortium, consisting of (1) nucleotide substitution per fourfold degenerate site in coding regions, (2) nucleotide substitution per site in relics of transposable elements active only before the human-mouse speciation, and (3) the nonaligning fraction of human DNA that is nonrepetitive or in ancestral repeats; and three derived from human genome data alone, consisting of (4) SNP density, (5) frequency of insertion of transposable elements, and (6) rate of recombination. Features 1 and 2 are measures of nucleotide substitutions at two classes of "neutral" sites, whereas 4 is a measure of recent mutations. Feature 3 is a measure dominated by deletions in mouse, whereas 5 represents insertions in human. It was found that all six vary significantly in megabase-sized regions genome-wide, and many vary together. This indicates that some regions of a genome change slowly by all processes that alter DNA, and others change faster. Regional variation in all processes is correlated with, but not completely accounted for, by GC content in human and the difference between GC content in human and mouse.
Figures





References
-
- Ansari-Lari M.A., Oeltjen, J.C., Schwartz, S., Zhang, Z., Muzny, D.M., Lu, J., Gorrell, J.H., Chinault, A.C., Belmont, J.W., Miller, W., et al. 1998. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8: 29-40. - PubMed
-
- Archibald J.D., Averianov, A.O., and Ekdale, E.G. 2001. Late Cretaceous relatives of rabbits, rodents, and other extant eutherian mammals. Nature 414: 62-65. - PubMed
-
- Begun D.J. and Aquadro, C.F. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519-520. - PubMed
-
- Bernardi G. 1986. Compositional constraints and genome evolution. J. Mol. Evol. 24: 1-11. - PubMed
-
- ___, 1993. The isochore organization of the human genome and its evolutionary history—A review. Gene 135: 57-66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
