Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map
- PMID: 12529305
- PMCID: PMC430948
- DOI: 10.1101/gr.830003
Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map
Abstract
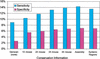
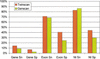
The availability of draft sequences for both the mouse and human genomes makes it possible, for the first time, to annotate whole mammalian genomes using comparative methods. TWINSCAN is a gene-prediction system that combines the methods of single-genome predictors like GENSCAN with information derived from genome comparison, thereby improving accuracy. Because TWINSCAN uses genomic sequence only, it is less biased toward highly and/or ubiquitously expressed genes than GENEWISE, GENOMESCAN, and other methods based on evidence derived from transcripts. We show that TWINSCAN improves gene prediction in human using intermediate products from various stages of the sequencing and analysis of the mouse genome, from low-redundancy, whole-genome shotgun reads to the draft assembly and the synteny map. TWINSCAN improves on the prior state of the art even when alignments from only 1X coverage of the mouse genome are available. Gene prediction accuracy improves steadily from 1X through 3X, more slowly from 3X to 4X, and relatively little thereafter. The assembly and the synteny map greatly speed the computations, however. Our human annotation using the mouse assembly is conservative, predicting only 25,622 genes, and appears to be one of the best de novo annotations of the human genome to date.
Figures






References
-
- Ansari-Lari M.A., Oeltjen, J.C., Schwartz, S., Zhang, Z., Muzny, D.M., Lu, J., Gorrell, J.H., Chinault, A.C., Belmont, J.W., Miller, W., et al. 1998. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 8: 29-40. - PubMed
-
- Ash R., 1965. Information theory. Wiley, New York.
-
- Bafna V. and Huson, D.H. 2000. The conserved exon method for gene finding. Proc. Int. Conf. Intell. Syst. Mol. Biol. 8: 3-12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
