Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ
- PMID: 12529404
- PMCID: PMC140696
- DOI: 10.1128/MCB.23.3.1004-1013.2003
Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ
Abstract
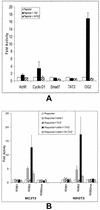
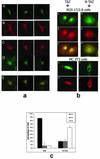
Core-binding factor 1 (Cbfa1; also called Runx2) is a transcription factor belonging to the Runt family of transcription factors that binds to an osteoblast-specific cis-acting element (OSE2) activating the expression of osteocalcin, an osteoblast-specific gene. Using the yeast two-hybrid system, we identified a transcriptional coactivator, TAZ (transcriptional coactivator with PDZ-binding motif), that binds to Cbfa1. A functional relationship between Cbfa1 and TAZ is demonstrated by the coimmunoprecipitation of TAZ by Cbfa1 and by the fact that TAZ induces a dose-dependent increase in the activity of osteocalcin promoter-luciferase constructs by Cbfa1. A dominant-negative construct of TAZ in which the coactivation domains have been deleted reduces osteocalcin gene expression down to basal levels. NIH 3T3, MC 3T3, and ROS 17/2.8 cells showed the expected nuclear localization of Cbfa1, whereas TAZ was distributed throughout the cytoplasm with some nuclear localization when transfected with either Cbfa1 or TAZ. Upon cotransfection by both Cbfa1 and TAZ, the transfected TAZ shows predominant nuclear localization. The dominant-negative construct of TAZ shows minimal nuclear localization upon cotransfection with Cbfa1. These data indicate that TAZ is a transcription coactivator for Cbfa1 and may be involved in the regulation of osteoblast differentiation.
Figures





References
-
- Aitken, A. 1996. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 6:341-347. - PubMed
-
- Albanese, C., J. Johnson, G. Watanabe, D. N. Eklund, Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21 mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. - PubMed
-
- Banerjee, C., L. R. McCabe, J.-Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Runt-homology domain proteins in osteoblast differentiation: AML-3/Cbfa1 is a major component of a bone specific complex. J. Cell. Biochem. 66:1-8. - PubMed
-
- Ducy, P. 2000. Cbfa1: a molecular switch in osteoblast biology. Dev. Dynam. 219:461-471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
