Global transcriptional responses of fission yeast to environmental stress
- PMID: 12529438
- PMCID: PMC140239
- DOI: 10.1091/mbc.e02-08-0499
Global transcriptional responses of fission yeast to environmental stress
Abstract
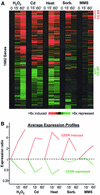
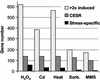
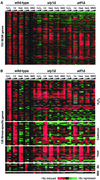
We explored transcriptional responses of the fission yeast Schizosaccharomyces pombe to various environmental stresses. DNA microarrays were used to characterize changes in expression profiles of all known and predicted genes in response to five stress conditions: oxidative stress caused by hydrogen peroxide, heavy metal stress caused by cadmium, heat shock caused by temperature increase to 39 degrees C, osmotic stress caused by sorbitol, and DNA damage caused by the alkylating agent methylmethane sulfonate. We define a core environmental stress response (CESR) common to all, or most, stresses. There was a substantial overlap between CESR genes of fission yeast and the genes of budding yeast that are stereotypically regulated during stress. CESR genes were controlled primarily by the stress-activated mitogen-activated protein kinase Sty1p and the transcription factor Atf1p. S. pombe also activated gene expression programs more specialized for a given stress or a subset of stresses. In general, these "stress-specific" responses were less dependent on the Sty1p mitogen-activated protein kinase pathway and may involve specific regulatory factors. Promoter motifs associated with some of the groups of coregulated genes were identified. We compare and contrast global regulation of stress genes in fission and budding yeasts and discuss evolutionary implications.
Figures



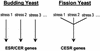
References
-
- Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol. 2000;71:129–134. - PubMed
-
- Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:244–250. - PubMed
-
- Cotto JJ, Morimoto RI. Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem Soc Symp. 1999;64:105–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

