Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics
- PMID: 12529507
- PMCID: PMC141043
- DOI: 10.1073/pnas.242716699
Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics
Abstract
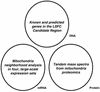
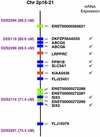
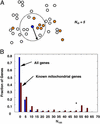
Identifying the genes responsible for human diseases requires combining information about gene position with clues about biological function. The recent availability of whole-genome data sets of RNA and protein expression provides powerful new sources of functional insight. Here we illustrate how such data sets can expedite disease-gene discovery, by using them to identify the gene causing Leigh syndrome, French-Canadian type (LSFC, Online Mendelian Inheritance in Man no. 220111), a human cytochrome c oxidase deficiency that maps to chromosome 2p16-21. Using four public RNA expression data sets, we assigned to all human genes a "score" reflecting their similarity in RNA-expression profiles to known mitochondrial genes. Using a large survey of organellar proteomics, we similarly classified human genes according to the likelihood of their protein product being associated with the mitochondrion. By intersecting this information with the relevant genomic region, we identified a single clear candidate gene, LRPPRC. Resequencing identified two mutations on two independent haplotypes, providing definitive genetic proof that LRPPRC indeed causes LSFC. LRPPRC encodes an mRNA-binding protein likely involved with mtDNA transcript processing, suggesting an additional mechanism of mitochondrial pathophysiology. Similar strategies to integrate diverse genomic information can be applied likewise to other disease pathways and will become increasingly powerful with the growing wealth of diverse, functional genomics data.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

