Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis
- PMID: 12529536
- PMCID: PMC166808
- DOI: 10.1104/pp.013029
Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis
Abstract
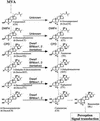
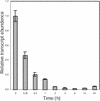
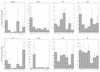
Brassinosteroids (BRs) are steroidal plant hormones that are essential for growth and development. There is only limited information on where BRs are synthesized and used. We studied the organ specificity of BR biosynthesis in Arabidopsis, using two different approaches: We analyzed the expression of BR-related genes using real-time quantitative reverse transcriptase-polymerase chain reaction, and analyzed endogenous BRs using gas chromatography-mass spectrometry. Before starting this study, we cloned the second BR-6-oxidase (BR6ox2) gene from Arabidopsis and found that the encoded enzyme has the same substrate specificity as the enzyme encoded by the previously isolated 6-oxidase gene (BR6ox1) of Arabidopsis. Endogenous BRs and the expression of BR-related genes were detected in all organs tested. The highest level of endogenous BRs and the highest expression of the BR6ox1, BR6ox2, and DWF4 genes were observed in apical shoots, which contain actively developing tissues. These genes are important in BR biosynthesis because they encode the rate-limiting or farthest downstream enzyme in the BR biosynthesis pathway. The second highest level of endogenous BRs and expression of BR6ox1 and DWF4 were observed in siliques, which contains actively developing embryos and seeds. These findings indicate that BRs are synthesized in all organs tested, but are most actively synthesized in young, actively developing organs. In contrast, synthesis was limited in mature organs. Our observations are consistent with the idea that BRs function as the growth-promoting hormone in plants.
Figures

References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem. 2001;276:25687–25691. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

