A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma
- PMID: 12531883
- PMCID: PMC151856
- DOI: 10.1172/JCI15109
A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma
Abstract
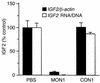
IGF-II is a mitogenic peptide that has been implicated in hepatocellular oncogenesis. Since the silencing of gene expression is frequently associated with cytosine methylation at cytosine-guanine (CpG) dinucleotides, we designed a methylated oligonucleotide (MON1) complementary to a region encompassing IGF2 promoter P4 in an attempt to induce DNA methylation at that locus and diminish IGF2 mRNA levels. MON1 specifically inhibited IGF2 mRNA accumulation in vitro, whereas an oligonucleotide (ON1) with the same sequence but with nonmethylated cytosines had no effect on IGF2 mRNA abundance. MON1 treatment led to the specific induction of de novo DNA methylation in the region of IGF2 promoter hP4. Cells from a human hepatocellular carcinoma (HCC) cell line, Hep 3B, were implanted into the livers of nude mice, resulting in the growth of large tumors. Animals treated with MON1 had markedly prolonged survival as compared with those animals treated with saline or a truncated methylated oligonucleotide that did not alter IGF2 mRNA levels in vitro. This study demonstrates that a methylated sense oligonucleotide can be used to induce epigenetic changes in the IGF2 gene and that inhibition of IGF2 mRNA accumulation may lead to enhanced survival in a model of HCC.
Figures
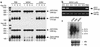





References
-
- Nakae J, Kido Y, Accili D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 2001;22:818–835. - PubMed
-
- Cariani E, et al. Differential expression of insulin-like growth factor II mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosis. Cancer Res. 1988;48:6844–6849. - PubMed
-
- Kim SO, Park JG, Lee YI. Increased expression of the insulin-like growth factor I (IGF-I) receptor gene in hepatocellular carcinoma cell lines: implications of IGF-I receptor gene activation by hepatitis B virus X gene product. Cancer Res. 1996;56:3831–3836. - PubMed
-
- Pulford DJ, Falls JG, Killian JK, Jirtle RL. Polymorphisms, genomic imprinting and cancer susceptibility. Mutat. Res. 1999;436:59–67. - PubMed
-
- Zhang L, et al. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

