An outer membrane enzyme that generates the 2-amino-2-deoxy-gluconate moiety of Rhizobium leguminosarum lipid A
- PMID: 12531907
- PMCID: PMC2745892
- DOI: 10.1074/jbc.M300378200
An outer membrane enzyme that generates the 2-amino-2-deoxy-gluconate moiety of Rhizobium leguminosarum lipid A
Abstract
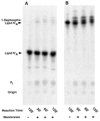
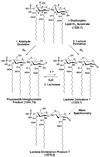
The structures of Rhizobium leguminosarum and Rhizobium etli lipid A are distinct from those found in other Gram-negative bacteria. Whereas the more typical Escherichia coli lipid A is a hexa-acylated disaccharide of glucosamine that is phosphorylated at positions 1 and 4', R. etli and R. leguminosarum lipid A consists of a mixture of structurally related species (designated A-E) that lack phosphate. A conserved distal unit, comprised of a diacylated glucosamine moiety with galacturonic acid residue at position 4' and a secondary 27-hydroxyoctacosanoyl (27-OH-C28) as part of a 2' acyloxyacyl moiety, is present in all five components. The proximal end is heterogeneous, differing in the number and lengths of acyl chains and in the identity of the sugar itself. A proximal glucosamine unit is present in B and C, but an unusual 2-amino-2-deoxy-gluconate moiety is found in D-1 and E. We now demonstrate that membranes of R. leguminosarum and R. etli can convert B to D-1 in a reaction that requires added detergent and is inhibited by EDTA. Membranes of Sinorhizobium meliloti and E. coli lack this activity. Mass spectrometry demonstrates that B is oxidized in vitro to a substance that is 16 atomic mass units larger, consistent with the formation of D-1. The oxidation of the lipid A proximal unit is also demonstrated by matrix-assisted laser desorption ionization time-of-flight mass spectrometry in the positive and negative modes using the model substrate, 1-dephospho-lipid IV(A). With this material, an additional intermediate (or by product) is detected that is tentatively identified as a lactone derivative of 1-dephospho-lipid IV(A). The enzyme, presumed to be an oxidase, is located exclusively in the outer membrane of R. leguminosarum as judged by sucrose gradient analysis. To our knowledge, an oxidase associated with the outer membranes of Gram-negative bacteria has not been reported previously.
Figures









Similar articles
-
Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane.Biochim Biophys Acta. 2008 Sep;1778(9):1881-96. doi: 10.1016/j.bbamem.2007.07.021. Epub 2007 Aug 11. Biochim Biophys Acta. 2008. PMID: 17880914 Free PMC article. Review.
-
Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ.J Biol Chem. 2003 Apr 4;278(14):12120-9. doi: 10.1074/jbc.M300379200. Epub 2003 Jan 15. J Biol Chem. 2003. PMID: 12531908 Free PMC article.
-
A deacylase in Rhizobium leguminosarum membranes that cleaves the 3-O-linked beta-hydroxymyristoyl moiety of lipid A precursors.J Biol Chem. 1999 Apr 16;274(16):11150-8. doi: 10.1074/jbc.274.16.11150. J Biol Chem. 1999. PMID: 10196200 Free PMC article.
-
Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion.J Biol Chem. 2000 Sep 8;275(36):28006-16. doi: 10.1074/jbc.M004008200. J Biol Chem. 2000. PMID: 10856303 Free PMC article.
-
A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium.J Biol Chem. 1996 Dec 13;271(50):32126-36. doi: 10.1074/jbc.271.50.32126. J Biol Chem. 1996. PMID: 8943266
Cited by
-
Lipoprotein PssN of Rhizobium leguminosarum bv. trifolii: subcellular localization and possible involvement in exopolysaccharide export.J Bacteriol. 2006 Oct;188(19):6943-52. doi: 10.1128/JB.00651-06. J Bacteriol. 2006. PMID: 16980497 Free PMC article.
-
The calcium-stimulated lipid A 3-O deacylase from Rhizobium etli is not essential for plant nodulation.Biochim Biophys Acta. 2012 Jul;1831(7):1250-9. doi: 10.1016/j.bbalip.2013.04.002. Epub 2013 Apr 12. Biochim Biophys Acta. 2012. PMID: 23583844 Free PMC article.
-
Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra.J Bacteriol. 2004 Oct;186(19):6617-25. doi: 10.1128/JB.186.19.6617-6625.2004. J Bacteriol. 2004. PMID: 15375143 Free PMC article.
-
Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A.J Biol Chem. 2004 Dec 31;279(53):55780-91. doi: 10.1074/jbc.M406480200. Epub 2004 Oct 15. J Biol Chem. 2004. PMID: 15489235 Free PMC article.
-
Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane.Biochim Biophys Acta. 2008 Sep;1778(9):1881-96. doi: 10.1016/j.bbamem.2007.07.021. Epub 2007 Aug 11. Biochim Biophys Acta. 2008. PMID: 17880914 Free PMC article. Review.
References
-
- Niner BM, Hirsch AM. Symbiosis. 1998;24:51–102.
-
- Stacey G, Burris RH, Evans HJ, editors. Biological Nitrogen Fixation. New York: Routledge, Chapman and Hall, Inc.; 1992.
-
- Triplett EW, editor. Prokaryotic Nitrogen Fixation: A Model System for the Analysis of a Biological Process. Wymondham, UK: Horizon Scientific Press; 2000.
-
- Downie JA, Walker SA. Curr. Opin. Plant Biol. 1999;2:483–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

