Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ
- PMID: 12531908
- PMCID: PMC2548327
- DOI: 10.1074/jbc.M300379200
Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ
Abstract
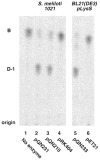
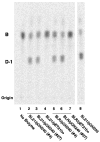
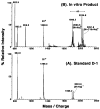
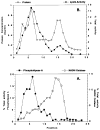
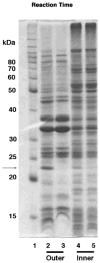
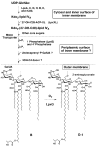
An unusual feature of the lipid A from the plant endosymbionts Rhizobium etli and Rhizobium leguminosarum is the presence of a proximal sugar unit consisting of a 2-amino-2-deoxy-gluconate moiety in place of glucosamine. An outer membrane oxidase that generates the 2-amino-2-deoxy-gluconate unit from a glucosamine-containing precursor is present in membranes of R. leguminosarum and R. etli but not in S. meliloti or Escherichia coli. We now report the identification of a hybrid cosmid that directs the overexpression of this activity by screening 1800 lysates of individual colonies of a R. leguminosarum 3841 genomic DNA library in the host strain R. etli CE3. Two cosmids (p1S11D and p1U12G) were identified in this manner and transferred into S. meliloti, in which they also directed the expression of oxidase activity in the absence of any chromosomal background. Subcloning and sequencing of the oxidase gene on a 6.5-kb fragment derived from the approximately 20-kb insert in p1S11D revealed that the enzyme is encoded by a gene (lpxQ) that specifies a protein of 224 amino acid residues with a putative signal sequence cleavage site at position 28. Heterologous expression of lpxQ using the T7lac promoter system in E. coli resulted in the production of catalytically active oxidase that was localized in the outer membrane. A new outer membrane protein of the size expected for LpxQ was present in this construct and was subjected to microsequencing to confirm its identity and the site of signal peptide cleavage. LpxQ expressed in E. coli generates the same products as seen in R. leguminosarum membranes. LpxQ is dependent on O(2) for activity, as demonstrated by inhibition of the reaction under strictly anaerobic conditions. An ortholog of LpxQ is present in the genome of Agrobacterium tumefaciens, as shown by heterologous expression of oxidase activity in E. coli.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

