Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene
- PMID: 12533452
- PMCID: PMC142808
- DOI: 10.1128/JB.185.3.772-778.2003
Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene
Abstract
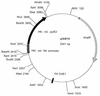
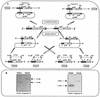
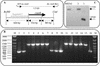
So far, the extremely halophilic archaeon Haloferax volcanii has the best genetic tools among the archaea. However, the lack of an efficient gene knockout system for this organism has hampered further genetic studies. In this paper we describe the development of pyrE-based positive selection and counterselection systems to generate an efficient gene knockout system. The H. volacanii pyrE1 and pyrE2 genes were isolated, and the pyrE2 gene was shown to code for the physiological enzyme orotate phosphoribosyl transferase. A DeltapyrE2 strain was constructed and used to isolate deletion mutants by the following two steps: (i) integration of a nonreplicative plasmid carrying both the pyrE2 wild-type gene, as a selectable marker, and a cloned chromosomal DNA fragment containing a deletion in the desired gene; and (ii) excision of the integrated plasmid after selection with 5-fluoroorotic acid. Application of this gene knockout system is described.
Figures

References
-
- Bell, S. D., and S. P. Jackson. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 6:222-228. - PubMed
-
- Bell, S. D., C. P. Magill, and S. P. Jackson. 2001. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 29:392-395. - PubMed
-
- Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. - PubMed
-
- Cline, S., F. Pfeifer, and W. Doolittle. 1995. Transformation of halophilic archaea, p. 197-204. In F. Robb (ed.), Protocols for archaeal research. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

