Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae
- PMID: 12533475
- PMCID: PMC142830
- DOI: 10.1128/JB.185.3.991-1000.2003
Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae
Abstract
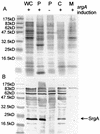
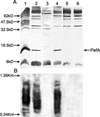
Disulfide oxidoreductases are viewed as foldases that help to maintain proteins on productive folding pathways by enhancing the rate of protein folding through the catalytic incorporation of disulfide bonds. SrgA, encoded on the virulence plasmid pStSR100 of Salmonella enterica serovar Typhimurium and located downstream of the plasmid-borne fimbrial operon, is a disulfide oxidoreductase. Sequence analysis indicates that SrgA is similar to DsbA from, for example, Escherichia coli, but not as highly conserved as most of the chromosomally encoded disulfide oxidoreductases from members of the family Enterobacteriaceae. SrgA is localized to the periplasm, and its disulfide oxidoreductase activity is dependent upon the presence of functional DsbB, the protein that is also responsible for reoxidation of the major disulfide oxidoreductase, DsbA. A quantitative analysis of the disulfide oxidoreductase activity of SrgA showed that SrgA was less efficient than DsbA at introducing disulfide bonds into the substrate alkaline phosphatase, suggesting that SrgA is more substrate specific than DsbA. It was also demonstrated that the disulfide oxidoreductase activity of SrgA is necessary for the production of plasmid-encoded fimbriae. The major structural subunit of the plasmid-encoded fimbriae, PefA, contains a disulfide bond that must be oxidized in order for PefA stability to be maintained and for plasmid-encoded fimbriae to be assembled. SrgA efficiently oxidizes the disulfide bond of PefA, while the S. enterica serovar Typhimurium chromosomally encoded disulfide oxidoreductase DsbA does not. pefA and srgA were also specifically expressed at pH 5.1 but not at pH 7.0, suggesting that the regulatory mechanisms involved in pef gene expression are also involved in srgA expression. SrgA therefore appears to be a substrate-specific disulfide oxidoreductase, thus explaining the requirement for an additional catalyst of disulfide bond formation in addition to DsbA of S. enterica serovar Typhimurium.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

